[English] 日本語
 Yorodumi
Yorodumi- EMDB-2976: Time-resolved Cryo Electron Microscopy of ribosome subunit association -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-2976 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Time-resolved Cryo Electron Microscopy of ribosome subunit association | |||||||||
 Map data Map data | Reconstruction of E. Coli naked 70S ribosome in non-rotated (NR) conformation | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | time-resolved / cryo-EM / mixing-spraying / ribosome subunit association / structural dynamics | |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 9.7 Å | |||||||||
 Authors Authors | Chen B / Kaledhonkar S / Sun M / Shen B / Lu Z / Barnard D / Lu T / Gonzalez Jr R / Frank J | |||||||||
 Citation Citation |  Journal: Structure / Year: 2015 Journal: Structure / Year: 2015Title: Structural dynamics of ribosome subunit association studied by mixing-spraying time-resolved cryogenic electron microscopy. Authors: Bo Chen / Sandip Kaledhonkar / Ming Sun / Bingxin Shen / Zonghuan Lu / David Barnard / Toh-Ming Lu / Ruben L Gonzalez / Joachim Frank /  Abstract: Ribosomal subunit association is a key checkpoint in translation initiation but its structural dynamics are poorly understood. Here, we used a recently developed mixing-spraying, time-resolved, ...Ribosomal subunit association is a key checkpoint in translation initiation but its structural dynamics are poorly understood. Here, we used a recently developed mixing-spraying, time-resolved, cryogenic electron microscopy (cryo-EM) method to study ribosomal subunit association in the sub-second time range. We have improved this method and increased the cryo-EM data yield by tenfold. Pre-equilibrium states of the association reaction were captured by reacting the mixture of ribosomal subunits for 60 ms and 140 ms. We also identified three distinct ribosome conformations in the associated ribosomes. The observed proportions of these conformations are the same in these two time points, suggesting that ribosomes equilibrate among the three conformations within less than 60 ms upon formation. Our results demonstrate that the mixing-spraying method can capture multiple states of macromolecules during a sub-second reaction. Other fast processes, such as translation initiation, decoding, and ribosome recycling, are amenable to study with this method. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_2976.map.gz emd_2976.map.gz | 14.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-2976-v30.xml emd-2976-v30.xml emd-2976.xml emd-2976.xml | 8.9 KB 8.9 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_2976.png emd_2976.png | 304.2 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-2976 http://ftp.pdbj.org/pub/emdb/structures/EMD-2976 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-2976 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-2976 | HTTPS FTP |
-Validation report
| Summary document |  emd_2976_validation.pdf.gz emd_2976_validation.pdf.gz | 243.6 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_2976_full_validation.pdf.gz emd_2976_full_validation.pdf.gz | 242.7 KB | Display | |
| Data in XML |  emd_2976_validation.xml.gz emd_2976_validation.xml.gz | 5 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-2976 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-2976 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-2976 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-2976 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_2976.map.gz / Format: CCP4 / Size: 15.3 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_2976.map.gz / Format: CCP4 / Size: 15.3 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Reconstruction of E. Coli naked 70S ribosome in non-rotated (NR) conformation | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
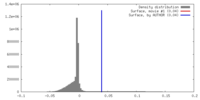
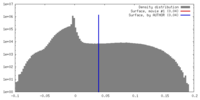
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : E. Coli 70S Ribosome
| Entire | Name: E. Coli 70S Ribosome |
|---|---|
| Components |
|
-Supramolecule #1000: E. Coli 70S Ribosome
| Supramolecule | Name: E. Coli 70S Ribosome / type: sample / ID: 1000 / Number unique components: 1 |
|---|
-Supramolecule #1: Escherichia coli 70S ribosome
| Supramolecule | Name: Escherichia coli 70S ribosome / type: complex / ID: 1 / Recombinant expression: No / Ribosome-details: ribosome-prokaryote: LSU 50S, SSU 30S |
|---|---|
| Source (natural) | Organism:  |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.6 Details: 25 mM Tris-HCl, 60 mM NH4Cl, 5 mM 2-mercaptoethanol, 3.5 mM MgCl2 |
|---|---|
| Grid | Details: Quantifoil R2/2 300 mesh copper grid with thin carbon sipport |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 80 % / Chamber temperature: 80 K / Instrument: OTHER Details: Equal volume of 1.2 microM 30S and 0.6 microM 50S (final concentration after mixing) were injected into the mixing-spraying device each at flow rate of 3 microL/s. The computer-controlled ...Details: Equal volume of 1.2 microM 30S and 0.6 microM 50S (final concentration after mixing) were injected into the mixing-spraying device each at flow rate of 3 microL/s. The computer-controlled plunging device was purchased from Dr. Howard White (Eastern Virginia Medical School, VA). Timed resolved state: Vitrified after spraying |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI F20 |
|---|---|
| Temperature | Average: 80 K |
| Details | Low dose, Data was collected over two years time |
| Date | Sep 13, 2013 |
| Image recording | Category: CCD / Film or detector model: GATAN ULTRASCAN 4000 (4k x 4k) / Number real images: 3402 / Average electron dose: 17 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated magnification: 66318 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2 mm / Nominal defocus max: 4.0 µm / Nominal defocus min: 2.0 µm / Nominal magnification: 50000 |
| Sample stage | Specimen holder model: GATAN LIQUID NITROGEN |
| Experimental equipment |  Model: Tecnai F20 / Image courtesy: FEI Company |
- Image processing
Image processing
| Details | The partciles were selected with Autopicker (Langlois et al., 2014), and 3D classification and reconstruction with RELION |
|---|---|
| CTF correction | Details: each Micrograph |
| Final reconstruction | Applied symmetry - Point group: C1 (asymmetric) / Resolution.type: BY AUTHOR / Resolution: 9.7 Å / Resolution method: OTHER / Software - Name: Arachnid, RELION, SPIDER / Number images used: 39678 |
 Movie
Movie Controller
Controller














 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)