[English] 日本語
 Yorodumi
Yorodumi- EMDB-25066: Structure of E. coli LetB delta (Ring6) mutant, Ring1 in the clos... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-25066 | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of E. coli LetB delta (Ring6) mutant, Ring1 in the closed state (Model 1) | |||||||||||||||
 Map data Map data | Full delta(Ring6) LetB map | |||||||||||||||
 Sample Sample |
| |||||||||||||||
 Keywords Keywords | lipid transport / bacterial cell envelope / MCE | |||||||||||||||
| Function / homology |  Function and homology information Function and homology informationintermembrane lipid transfer / membrane organization / outer membrane-bounded periplasmic space / identical protein binding / plasma membrane Similarity search - Function | |||||||||||||||
| Biological species |  | |||||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.2 Å | |||||||||||||||
 Authors Authors | Vieni C / Coudray N | |||||||||||||||
| Funding support |  United States, 4 items United States, 4 items
| |||||||||||||||
 Citation Citation |  Journal: J Mol Biol / Year: 2022 Journal: J Mol Biol / Year: 2022Title: Role of Ring6 in the Function of the E. coli MCE Protein LetB. Authors: Casey Vieni / Nicolas Coudray / Georgia L Isom / Gira Bhabha / Damian C Ekiert /  Abstract: LetB is a tunnel-forming protein found in the cell envelope of some double-membraned bacteria, and is thought to be important for the transport of lipids between the inner and outer membranes. In ...LetB is a tunnel-forming protein found in the cell envelope of some double-membraned bacteria, and is thought to be important for the transport of lipids between the inner and outer membranes. In Escherichia coli the LetB tunnel is formed from a stack of seven rings (Ring1 - Ring7), in which each ring is composed of a homo-hexameric assembly of MCE domains. The primary sequence of each MCE domain of the LetB protein is substantially divergent from the others, making each MCE ring unique in nature. The role of each MCE domain and how it contributes to the function of LetB is not well understood. Here we probed the importance of each MCE ring for the function of LetB, using a combination of bacterial growth assays and cryo-EM. Surprisingly, we find that ΔRing3 and ΔRing6 mutants, in which Ring3 and Ring6 have been deleted, confer increased resistance to membrane perturbing agents. Specific mutations in the pore-lining loops of Ring6 similarly confer increased resistance. A cryo-EM structure of the ΔRing6 mutant shows that despite the absence of Ring6, which leads to a shorter assembly, the overall architecture is maintained, highlighting the modular nature of MCE proteins. Previous work has shown that Ring6 is dynamic and in its closed state, may restrict the passage of substrate through the tunnel. Our work suggests that removal of Ring6 may relieve this restriction. The deletion of Ring6 combined with mutations in the pore-lining loops leads to a model for the tunnel gating mechanism of LetB. Together, these results provide insight into the functional roles of individual MCE domains and pore-lining loops in the LetB protein. | |||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_25066.map.gz emd_25066.map.gz | 192.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-25066-v30.xml emd-25066-v30.xml emd-25066.xml emd-25066.xml | 22.9 KB 22.9 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_25066_fsc.xml emd_25066_fsc.xml | 14.1 KB | Display |  FSC data file FSC data file |
| Images |  emd_25066.png emd_25066.png | 100 KB | ||
| Masks |  emd_25066_msk_1.map emd_25066_msk_1.map | 244.1 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-25066.cif.gz emd-25066.cif.gz | 6.8 KB | ||
| Others |  emd_25066_additional_1.map.gz emd_25066_additional_1.map.gz emd_25066_half_map_1.map.gz emd_25066_half_map_1.map.gz emd_25066_half_map_2.map.gz emd_25066_half_map_2.map.gz | 225.3 MB 193.3 MB 193.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-25066 http://ftp.pdbj.org/pub/emdb/structures/EMD-25066 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-25066 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-25066 | HTTPS FTP |
-Validation report
| Summary document |  emd_25066_validation.pdf.gz emd_25066_validation.pdf.gz | 779.7 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_25066_full_validation.pdf.gz emd_25066_full_validation.pdf.gz | 779.3 KB | Display | |
| Data in XML |  emd_25066_validation.xml.gz emd_25066_validation.xml.gz | 21.3 KB | Display | |
| Data in CIF |  emd_25066_validation.cif.gz emd_25066_validation.cif.gz | 28.3 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-25066 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-25066 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-25066 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-25066 | HTTPS FTP |
-Related structure data
| Related structure data |  7seeMC  7sefC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | |
| EM raw data |  EMPIAR-10821 (Title: Cryo EM structure of ΔRing6 LetB / Data size: 3.0 TB EMPIAR-10821 (Title: Cryo EM structure of ΔRing6 LetB / Data size: 3.0 TBData #1: Unaligned multi-frame micrographs of delta(Ring6) LetB [micrographs - multiframe]) |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_25066.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_25066.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Full delta(Ring6) LetB map | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.272 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
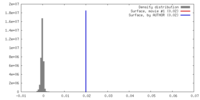
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_25066_msk_1.map emd_25066_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
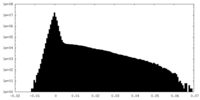
| Density Histograms |
-Additional map: Post processed map from RELION
| File | emd_25066_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Post processed map from RELION | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half Map #1
| File | emd_25066_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half Map #1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half Map #2
| File | emd_25066_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half Map #2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Homohexameric complex of delta(Ring6) mutant form of LetB
| Entire | Name: Homohexameric complex of delta(Ring6) mutant form of LetB |
|---|---|
| Components |
|
-Supramolecule #1: Homohexameric complex of delta(Ring6) mutant form of LetB
| Supramolecule | Name: Homohexameric complex of delta(Ring6) mutant form of LetB type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 473.25 KDa |
-Macromolecule #1: MCE family protein, Intermembrane transport protein YebT chimera
| Macromolecule | Name: MCE family protein, Intermembrane transport protein YebT chimera type: protein_or_peptide / ID: 1 / Number of copies: 6 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 78.967773 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MQDRGNTVTI DFMSADGIVP GRTPVRYQGV EVGTVQDISL SDDLRKIEVK VSIKSDMKDA LREETQFWLV TPKASLAGVS GLDALVGGN YIGMMPGKGK EQDHFVALDT QPKYRLDNGD LMIHLQAPDL GSLNSGSLVY FRKIPVGKVY DYAINPNKQG V VIDVLIER ...String: MQDRGNTVTI DFMSADGIVP GRTPVRYQGV EVGTVQDISL SDDLRKIEVK VSIKSDMKDA LREETQFWLV TPKASLAGVS GLDALVGGN YIGMMPGKGK EQDHFVALDT QPKYRLDNGD LMIHLQAPDL GSLNSGSLVY FRKIPVGKVY DYAINPNKQG V VIDVLIER RFTDLVKKGS RFWNVSGVDA NVSISGAKVK LESLAALVNG AIAFDSPEES KPAEAEDTFG LYEDLAHSQR GV IIKLELP SGAGLTADST PLMYQGLEVG QLTKLDLNPG GKVTGEMTVD PSVVTLLREN TRIELRNPKL SLSDANLSAL LTG KTFELV PGDGEPRKEF VVVPGEKALL HEPDVLTLTL TAPESYGIDA GQPLILHGVQ VGQVIDRKLT SKGVTFTVAI EPQH RELVK GDSKFVVNSR VDVKVGLDGV EFLGASASEW INGGIRILPG DKGEMKASYP LYANLEKALE NSLSDLPTTT VSLSA ETLP DVQAGSVVLY RKFEVGEVIT VRPRANAFDI DLHIKPEYRN LLTSNSVFWA EGGAKVQLNG SGLTVQASPL SRALKG AIS FDNLSGASAS QRKGDKRILY ASETAARAVG LSIIVEAPEA GSLGIGTPVL FRGLEVGTVT GMTLGTLSDR VMIAMRI SK RYQHLVRNNS VFWLASGYSL DFGLTGGVVK TGTFNQFIRG GIAFATPPGT PLAPKAQEGK HFLLQESEPK EWREWGTA L PKGETHHHHH H UniProtKB: UNIPROTKB: A0A769F599, Intermembrane transport protein YebT |
-Macromolecule #2: UNKNOWN ATOM OR ION
| Macromolecule | Name: UNKNOWN ATOM OR ION / type: ligand / ID: 2 / Number of copies: 6 / Formula: UNX |
|---|
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.75 mg/mL | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 8 Component:
Details: This buffer was used as gel filtration buffer | |||||||||
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY / Support film - Film thickness: 120 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 12 sec. | |||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 96 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV Details: 3 second blot time and blot force of 5 before plunge freezing. | |||||||||
| Details | This sample was monodisperse |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Digitization - Dimensions - Width: 11520 pixel / Digitization - Dimensions - Height: 8184 pixel / Number grids imaged: 1 / Number real images: 10763 / Average exposure time: 1.5 sec. / Average electron dose: 56.3 e/Å2 Details: Images were collected in super resolution mode with a pixel size of 0.318A |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.2 µm / Nominal defocus min: 0.8 µm / Nominal magnification: 37000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model |
| ||||||
|---|---|---|---|---|---|---|---|
| Refinement | Space: REAL / Protocol: FLEXIBLE FIT | ||||||
| Output model |  PDB-7see: |
 Movie
Movie Controller
Controller









 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)