[English] 日本語
 Yorodumi
Yorodumi- EMDB-22511: Cryo-EM structure of Bromocriptine-bound dopamine receptor 2 in c... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-22511 | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of Bromocriptine-bound dopamine receptor 2 in complex with Gi protein | |||||||||||||||
 Map data Map data | ||||||||||||||||
 Sample Sample |
| |||||||||||||||
 Keywords Keywords | Dopamine receptor 2 / Gi protein / bromocriptine / SIGNALING PROTEIN | |||||||||||||||
| Function / homology |  Function and homology information Function and homology informationnegative regulation of dopamine receptor signaling pathway / positive regulation of dopamine uptake involved in synaptic transmission / negative regulation of dephosphorylation / positive regulation of glial cell-derived neurotrophic factor production / acid secretion / dopamine neurotransmitter receptor activity, coupled via Gi/Go / nervous system process involved in regulation of systemic arterial blood pressure / response to histamine / negative regulation of circadian sleep/wake cycle, sleep / regulation of synapse structural plasticity ...negative regulation of dopamine receptor signaling pathway / positive regulation of dopamine uptake involved in synaptic transmission / negative regulation of dephosphorylation / positive regulation of glial cell-derived neurotrophic factor production / acid secretion / dopamine neurotransmitter receptor activity, coupled via Gi/Go / nervous system process involved in regulation of systemic arterial blood pressure / response to histamine / negative regulation of circadian sleep/wake cycle, sleep / regulation of synapse structural plasticity / regulation of locomotion involved in locomotory behavior / neuron-neuron synaptic transmission / adenohypophysis development / negative regulation of dopamine secretion / negative regulation of cellular response to hypoxia / hyaloid vascular plexus regression / adenylate cyclase-inhibiting dopamine receptor signaling pathway / cerebral cortex GABAergic interneuron migration / positive regulation of renal sodium excretion / response to inactivity / orbitofrontal cortex development / regulation of potassium ion transport / Dopamine receptors / negative regulation of neuron migration / dopamine binding / branching morphogenesis of a nerve / regulation of dopamine uptake involved in synaptic transmission / positive regulation of growth hormone secretion / phospholipase C-activating dopamine receptor signaling pathway / peristalsis / heterotrimeric G-protein binding / drinking behavior / G protein-coupled receptor complex / grooming behavior / behavioral response to ethanol / auditory behavior / striatum development / positive regulation of G protein-coupled receptor signaling pathway / dopaminergic synapse / positive regulation of urine volume / positive regulation of multicellular organism growth / G protein-coupled receptor internalization / non-motile cilium / heterocyclic compound binding / negative regulation of synaptic transmission, glutamatergic / response to iron ion / adult walking behavior / arachidonate secretion / ciliary membrane / response to morphine / negative regulation of cytosolic calcium ion concentration / temperature homeostasis / positive regulation of neuroblast proliferation / regulation of synaptic transmission, GABAergic / positive regulation of cytokinesis / pigmentation / dopamine metabolic process / dopamine uptake involved in synaptic transmission / regulation of dopamine secretion / response to light stimulus / cellular response to ethanol / positive regulation of receptor internalization / associative learning / lateral plasma membrane / neuroblast proliferation / G-protein alpha-subunit binding / endocytic vesicle / negative regulation of protein secretion / potassium channel regulator activity / long-term memory / prepulse inhibition / response to axon injury / sperm flagellum / postsynaptic modulation of chemical synaptic transmission / adenylate cyclase inhibitor activity / synapse assembly / negative regulation of blood pressure / regulation of sodium ion transport / behavioral response to cocaine / positive regulation of protein localization to cell cortex / cellular response to retinoic acid / T cell migration / Adenylate cyclase inhibitory pathway / D2 dopamine receptor binding / response to prostaglandin E / release of sequestered calcium ion into cytosol / axon terminus / ionotropic glutamate receptor binding / G protein-coupled serotonin receptor binding / adenylate cyclase regulator activity / adenylate cyclase-inhibiting serotonin receptor signaling pathway / presynaptic modulation of chemical synaptic transmission / acrosomal vesicle / axonogenesis / negative regulation of phosphatidylinositol 3-kinase/protein kinase B signal transduction / cellular response to forskolin / regulation of heart rate / negative regulation of innate immune response / negative regulation of cell migration / regulation of mitotic spindle organization Similarity search - Function | |||||||||||||||
| Biological species |  Homo sapiens (human) / synthetic construct (others) Homo sapiens (human) / synthetic construct (others) | |||||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.8 Å | |||||||||||||||
 Authors Authors | Zhuang Y / Xu P | |||||||||||||||
| Funding support |  China, China,  United States, 4 items United States, 4 items
| |||||||||||||||
 Citation Citation |  Journal: Cell / Year: 2021 Journal: Cell / Year: 2021Title: Structural insights into the human D1 and D2 dopamine receptor signaling complexes. Authors: Youwen Zhuang / Peiyu Xu / Chunyou Mao / Lei Wang / Brian Krumm / X Edward Zhou / Sijie Huang / Heng Liu / Xi Cheng / Xi-Ping Huang / Dan-Dan Shen / Tinghai Xu / Yong-Feng Liu / Yue Wang / ...Authors: Youwen Zhuang / Peiyu Xu / Chunyou Mao / Lei Wang / Brian Krumm / X Edward Zhou / Sijie Huang / Heng Liu / Xi Cheng / Xi-Ping Huang / Dan-Dan Shen / Tinghai Xu / Yong-Feng Liu / Yue Wang / Jia Guo / Yi Jiang / Hualiang Jiang / Karsten Melcher / Bryan L Roth / Yan Zhang / Cheng Zhang / H Eric Xu /   Abstract: The D1- and D2-dopamine receptors (D1R and D2R), which signal through G and G, respectively, represent the principal stimulatory and inhibitory dopamine receptors in the central nervous system. D1R ...The D1- and D2-dopamine receptors (D1R and D2R), which signal through G and G, respectively, represent the principal stimulatory and inhibitory dopamine receptors in the central nervous system. D1R and D2R also represent the main therapeutic targets for Parkinson's disease, schizophrenia, and many other neuropsychiatric disorders, and insight into their signaling is essential for understanding both therapeutic and side effects of dopaminergic drugs. Here, we report four cryoelectron microscopy (cryo-EM) structures of D1R-G and D2R-G signaling complexes with selective and non-selective dopamine agonists, including two currently used anti-Parkinson's disease drugs, apomorphine and bromocriptine. These structures, together with mutagenesis studies, reveal the conserved binding mode of dopamine agonists, the unique pocket topology underlying ligand selectivity, the conformational changes in receptor activation, and potential structural determinants for G protein-coupling selectivity. These results provide both a molecular understanding of dopamine signaling and multiple structural templates for drug design targeting the dopaminergic system. | |||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_22511.map.gz emd_22511.map.gz | 26.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-22511-v30.xml emd-22511-v30.xml emd-22511.xml emd-22511.xml | 20.8 KB 20.8 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_22511.png emd_22511.png | 63.8 KB | ||
| Filedesc metadata |  emd-22511.cif.gz emd-22511.cif.gz | 7.3 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-22511 http://ftp.pdbj.org/pub/emdb/structures/EMD-22511 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-22511 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-22511 | HTTPS FTP |
-Related structure data
| Related structure data |  7jvrMC  7jv5C  7jvpC  7jvqC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_22511.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_22511.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.045 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Bromocriptin-bound dopamine receptor 2 in complex with Gi protein
| Entire | Name: Bromocriptin-bound dopamine receptor 2 in complex with Gi protein |
|---|---|
| Components |
|
-Supramolecule #1: Bromocriptin-bound dopamine receptor 2 in complex with Gi protein
| Supramolecule | Name: Bromocriptin-bound dopamine receptor 2 in complex with Gi protein type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#5 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Soluble cytochrome b562,D(2) dopamine receptor
| Macromolecule | Name: Soluble cytochrome b562,D(2) dopamine receptor / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 67.234578 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: DYKDDDDAKL QTMHHHHHHH HHHHHHHHAD LEDNWETLND NLKVIEKADN AAQVKDALTK MRAAALDAQK ATPPKLEDKS PDSPEMKDF RHGFDILVGQ IDDALKLANE GKVKEAQAAA EQLKTTRNAY IQKYLASENL YFQGGTMDPL NLSWYDDDLE R QNWSRPFN ...String: DYKDDDDAKL QTMHHHHHHH HHHHHHHHAD LEDNWETLND NLKVIEKADN AAQVKDALTK MRAAALDAQK ATPPKLEDKS PDSPEMKDF RHGFDILVGQ IDDALKLANE GKVKEAQAAA EQLKTTRNAY IQKYLASENL YFQGGTMDPL NLSWYDDDLE R QNWSRPFN GSDGKADRPH YNYYATLLTL LIAVIVFGNV LVCMAVSREK ALQTTTNYLI VSLAVADLLV ATLVMPWVVY LE VVGEWKF SRIHCDIFVT LDVMMCTASI LNLCAISIDR YTAVAMPMLY NTRYSSKRRV TVMISIVWVL SFTISCPLLF GLN NADQNE CIIANPAFVV YSSIVSFYVP FIVTLLVYIK IYIVLRRRRK RVNTKRSSRA FRAHLRAPLK GNCTHPEDMK LCTV IMKSN GSFPVNRRRV EAARRAQELE MEMLSSTSPP ERTRYSPIPP SHHQLTLPDP SHHGLHSTPD SPAKPEKNGH AKDHP KIAK IFEIQTMPNG KTRTSLKTMS RRKLSQQKEK KATQMLAIVL GVFIICWLPF FITHILNIHC DCNIPPVLYS AFTWLG YVN SAVNPIIYTT FNIEFRKAFL KILHC UniProtKB: Soluble cytochrome b562, D(2) dopamine receptor |
-Macromolecule #2: Guanine nucleotide-binding protein G(i) subunit alpha-1
| Macromolecule | Name: Guanine nucleotide-binding protein G(i) subunit alpha-1 type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 40.414047 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MGCTLSAEDK AAVERSKMID RNLREDGEKA AREVKLLLLG AGESGKNTIV KQMKIIHEAG YSEEECKQYK AVVYSNTIQS IIAIIRAMG RLKIDFGDSA RADDARQLFV LAGAAEEGFM TAELAGVIKR LWKDSGVQAC FNRSREYQLN DSAAYYLNDL D RIAQPNYI ...String: MGCTLSAEDK AAVERSKMID RNLREDGEKA AREVKLLLLG AGESGKNTIV KQMKIIHEAG YSEEECKQYK AVVYSNTIQS IIAIIRAMG RLKIDFGDSA RADDARQLFV LAGAAEEGFM TAELAGVIKR LWKDSGVQAC FNRSREYQLN DSAAYYLNDL D RIAQPNYI PTQQDVLRTR VKTTGIVETH FTFKDLHFKM FDVGAQRSER KKWIHCFEGV TAIIFCVALS DYDLVLAEDE EM NRMHASM KLFDSICNNK WFTDTSIILF LNKKDLFEEK IKKSPLTICY PEYAGSNTYE EAAAYIQCQF EDLNKRKDTK EIY THFTCS TDTKNVQFVF DAVTDVIIKN NLKDCGLF UniProtKB: Guanine nucleotide-binding protein G(i) subunit alpha-1 |
-Macromolecule #3: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1
| Macromolecule | Name: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 38.146707 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MGSLLQSELD QLRQEAEQLK NQIRDARKAC ADATLSQITN NIDPVGRIQM RTRRTLRGHL AKIYAMHWGT DSRLLVSASQ DGKLIIWDS YTTNKVHAIP LRSSWVMTCA YAPSGNYVAC GGLDNICSIY NLKTREGNVR VSRELAGHTG YLSCCRFLDD N QIVTSSGD ...String: MGSLLQSELD QLRQEAEQLK NQIRDARKAC ADATLSQITN NIDPVGRIQM RTRRTLRGHL AKIYAMHWGT DSRLLVSASQ DGKLIIWDS YTTNKVHAIP LRSSWVMTCA YAPSGNYVAC GGLDNICSIY NLKTREGNVR VSRELAGHTG YLSCCRFLDD N QIVTSSGD TTCALWDIET GQQTTTFTGH TGDVMSLSLA PDTRLFVSGA CDASAKLWDV REGMCRQTFT GHESDINAIC FF PNGNAFA TGSDDATCRL FDLRADQELM TYSHDNIICG ITSVSFSKSG RLLLAGYDDF NCNVWDALKA DRAGVLAGHD NRV SCLGVT DDGMAVATGS WDSFLKIWNG SS UniProtKB: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 |
-Macromolecule #4: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2
| Macromolecule | Name: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2 type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 7.861143 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MASNNTASIA QARKLVEQLK MEANIDRIKV SKAAADLMAY CEAHAKEDPL LTPVPASENP FREKKFFCAI L UniProtKB: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2 |
-Macromolecule #5: Antibody fragment ScFv16
| Macromolecule | Name: Antibody fragment ScFv16 / type: protein_or_peptide / ID: 5 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 28.813047 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: DVQLVESGGG LVQPGGSRKL SCSASGFAFS SFGMHWVRQA PEKGLEWVAY ISSGSGTIYY ADTVKGRFTI SRDDPKNTLF LQMTSLRSE DTAMYYCVRS IYYYGSSPFD FWGQGTTLTV SSGGGGSGGG GSGGGGSDIV MTQATSSVPV TPGESVSISC R SSKSLLHS ...String: DVQLVESGGG LVQPGGSRKL SCSASGFAFS SFGMHWVRQA PEKGLEWVAY ISSGSGTIYY ADTVKGRFTI SRDDPKNTLF LQMTSLRSE DTAMYYCVRS IYYYGSSPFD FWGQGTTLTV SSGGGGSGGG GSGGGGSDIV MTQATSSVPV TPGESVSISC R SSKSLLHS NGNTYLYWFL QRPGQSPQLL IYRMSNLASG VPDRFSGSGS GTAFTLTISR LEAEDVGVYY CMQHLEYPLT FG AGTKLEL KGSLEVLFQG PAAAHHHHHH HH |
-Macromolecule #6: bromoergocryptine
| Macromolecule | Name: bromoergocryptine / type: ligand / ID: 6 / Number of copies: 1 / Formula: 08Y |
|---|---|
| Molecular weight | Theoretical: 654.594 Da |
| Chemical component information | 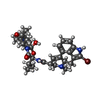 ChemComp-08Y: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.2 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 BASE (4k x 4k) / Average electron dose: 64.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller















































 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)






















