+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-22386 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of chicken CLC-7 | |||||||||
 Map data Map data | Density map of chicken CLC-7 | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Lysosomal / chloride-proton antiporter / chloride transport / ion transport / proton transport / MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationStimuli-sensing channels / voltage-gated chloride channel activity / chloride transmembrane transporter activity / transmembrane transporter activity / membrane => GO:0016020 / intracellular membrane-bounded organelle / nucleoplasm / membrane Similarity search - Function | |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.92 Å | |||||||||
 Authors Authors | Schrecker M / Hite R | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: Elife / Year: 2020 Journal: Elife / Year: 2020Title: Cryo-EM structure of the lysosomal chloride-proton exchanger CLC-7 in complex with OSTM1. Authors: Marina Schrecker / Julia Korobenko / Richard K Hite /  Abstract: The chloride-proton exchanger CLC-7 plays critical roles in lysosomal homeostasis and bone regeneration and its mutation can lead to osteopetrosis, lysosomal storage disease and neurological ...The chloride-proton exchanger CLC-7 plays critical roles in lysosomal homeostasis and bone regeneration and its mutation can lead to osteopetrosis, lysosomal storage disease and neurological disorders. In lysosomes and the ruffled border of osteoclasts, CLC-7 requires a β-subunit, OSTM1, for stability and activity. Here, we present electron cryomicroscopy structures of CLC-7 in occluded states by itself and in complex with OSTM1, determined at resolutions up to 2.8 Å. In the complex, the luminal surface of CLC-7 is entirely covered by a dimer of the heavily glycosylated and disulfide-bonded OSTM1, which serves to protect CLC-7 from the degradative environment of the lysosomal lumen. OSTM1 binding does not induce large-scale rearrangements of CLC-7, but does have minor effects on the conformation of the ion-conduction pathway, potentially contributing to its regulatory role. These studies provide insights into the role of OSTM1 and serve as a foundation for understanding the mechanisms of CLC-7 regulation. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_22386.map.gz emd_22386.map.gz | 200.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-22386-v30.xml emd-22386-v30.xml emd-22386.xml emd-22386.xml | 14.7 KB 14.7 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_22386.png emd_22386.png | 182.6 KB | ||
| Filedesc metadata |  emd-22386.cif.gz emd-22386.cif.gz | 6.2 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-22386 http://ftp.pdbj.org/pub/emdb/structures/EMD-22386 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-22386 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-22386 | HTTPS FTP |
-Validation report
| Summary document |  emd_22386_validation.pdf.gz emd_22386_validation.pdf.gz | 389.6 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_22386_full_validation.pdf.gz emd_22386_full_validation.pdf.gz | 389.1 KB | Display | |
| Data in XML |  emd_22386_validation.xml.gz emd_22386_validation.xml.gz | 7.2 KB | Display | |
| Data in CIF |  emd_22386_validation.cif.gz emd_22386_validation.cif.gz | 8.2 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-22386 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-22386 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-22386 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-22386 | HTTPS FTP |
-Related structure data
| Related structure data |  7jm6MC  7jm7C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_22386.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_22386.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Density map of chicken CLC-7 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.71487 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Chicken CLC-7
| Entire | Name: Chicken CLC-7 |
|---|---|
| Components |
|
-Supramolecule #1: Chicken CLC-7
| Supramolecule | Name: Chicken CLC-7 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Chloride channel protein
| Macromolecule | Name: Chloride channel protein / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 88.684609 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MANVAKKVSW SGRDPRDDED ERAGETTPLL NGTGPGSAGG ARQFTPSSFL RPGQLSNVDL NEDIRELETE LPRPYPNEIP HNEKLLSLK YESLDYDNSE NQLFLEEERR INHAAFRTVE IKRWVICAMI GILTGLVACF IDIVVENLAG LKYRVVKDNI D KFTEKGGL ...String: MANVAKKVSW SGRDPRDDED ERAGETTPLL NGTGPGSAGG ARQFTPSSFL RPGQLSNVDL NEDIRELETE LPRPYPNEIP HNEKLLSLK YESLDYDNSE NQLFLEEERR INHAAFRTVE IKRWVICAMI GILTGLVACF IDIVVENLAG LKYRVVKDNI D KFTEKGGL SFSLLLWATL NASVVMVGSV IVAFIEPVAA GSGIPQIKCY LNGVKIPHVV RLKTLVIKVC GVILSVVGGL AV GKEGPMI HSGAVIAAGI SQGRSTSLKR DFKIFEYFRR DTEKRDFVSA GAAAGVSAAF GAPVGGVLFS LEEGASFWNQ FLT WRIFFA SMISTFTLNS VLSVYHGNAW DLSSPGLINF GRFDSEKMGY TIQEIPIFIF MGVVGGILGA LFNALNYWLT MFRI RYIHR PCLQVIEAML VAAVTAAVGF VMIYCSRDCQ PIQGSSVAYP LQLFCADGEY NSMATAFFNT PEKSVVNLFH DPPGS YNPM TLGMFTLMYF FLACWTYGLT VSAGVFIPSL LIGAAWGRLF GISLSYLSKG SIWADPGKYA LMGAAAQLGG IVRMTL SLT VIMMEATGNV TYGFPIMLVL MTAKIVGDYF VEGLYDMHIQ LQSVPFLHWE APVTSHSLTA REVMSTPVTC LRRIERV GT VVDILSDTSS NHNGFPVVES NPNTTQVAGL RGLILRSQLI VLLKHKVFVE RANLNLVQRR LKLKDFRDAY PRFPPIQS I HVSQDERECM IDLSEFMNPS PYTVPREASL PRVFKLFRAL GLRHLVVVNN HNEVVGMVTR KDLARYRLGK EGLEELSLA QT UniProtKB: Chloride channel protein |
-Macromolecule #2: ADENOSINE-5'-TRIPHOSPHATE
| Macromolecule | Name: ADENOSINE-5'-TRIPHOSPHATE / type: ligand / ID: 2 / Number of copies: 2 / Formula: ATP |
|---|---|
| Molecular weight | Theoretical: 507.181 Da |
| Chemical component information |  ChemComp-ATP: |
-Macromolecule #3: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 3 / Number of copies: 2 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
-Macromolecule #4: CHLORIDE ION
| Macromolecule | Name: CHLORIDE ION / type: ligand / ID: 4 / Number of copies: 6 / Formula: CL |
|---|---|
| Molecular weight | Theoretical: 35.453 Da |
-Macromolecule #5: (2R)-3-{[(R)-hydroxy{[(1S,2R,3S,4S,5R,6R)-2,3,4,6-tetrahydroxy-5-...
| Macromolecule | Name: (2R)-3-{[(R)-hydroxy{[(1S,2R,3S,4S,5R,6R)-2,3,4,6-tetrahydroxy-5-(phosphonooxy)cyclohexyl]oxy}phosphoryl]oxy}propane-1,2-diyl dinonanoate type: ligand / ID: 5 / Number of copies: 2 / Formula: 0J1 |
|---|---|
| Molecular weight | Theoretical: 694.64 Da |
| Chemical component information | 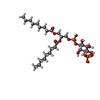 ChemComp-0J1: |
-Macromolecule #6: water
| Macromolecule | Name: water / type: ligand / ID: 6 / Number of copies: 72 / Formula: HOH |
|---|---|
| Molecular weight | Theoretical: 18.015 Da |
| Chemical component information |  ChemComp-HOH: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 2 mg/mL | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 8 Component:
| ||||||||||
| Grid | Model: Quantifoil R1.2/1.3 / Material: GOLD / Mesh: 400 / Support film - Material: CARBON / Support film - topology: HOLEY ARRAY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 10 sec. | ||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 295 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: SUPER-RESOLUTION / Average exposure time: 8.0 sec. / Average electron dose: 44.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: OTHER / Cs: 2.7 mm / Nominal defocus max: -2.5 µm / Nominal defocus min: -1.2 µm / Nominal magnification: 22500 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: OTHER |
|---|---|
| Output model |  PDB-7jm6: |
 Movie
Movie Controller
Controller
















 Z (Sec.)
Z (Sec.) X (Row.)
X (Row.) Y (Col.)
Y (Col.)





















