+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-21532 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
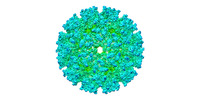
| Title | Native Mayaro virus cryo EM map, resolution 4.8 Angstroms | |||||||||
 Map data Map data | masked not sharpened | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | virus | |||||||||
| Function / homology |  Function and homology information Function and homology informationtogavirin / T=4 icosahedral viral capsid / symbiont-mediated suppression of host toll-like receptor signaling pathway / host cell cytoplasm / symbiont entry into host cell / serine-type endopeptidase activity / fusion of virus membrane with host endosome membrane / host cell nucleus / virion attachment to host cell / host cell plasma membrane ...togavirin / T=4 icosahedral viral capsid / symbiont-mediated suppression of host toll-like receptor signaling pathway / host cell cytoplasm / symbiont entry into host cell / serine-type endopeptidase activity / fusion of virus membrane with host endosome membrane / host cell nucleus / virion attachment to host cell / host cell plasma membrane / structural molecule activity / virion membrane / proteolysis / RNA binding / membrane Similarity search - Function | |||||||||
| Biological species |  Mayaro virus / Mayaro virus /  Mayaro virus (strain Brazil) Mayaro virus (strain Brazil) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 4.8 Å | |||||||||
 Authors Authors | Miller AS / Kuhn RJ | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Cell Host Microbe / Year: 2020 Journal: Cell Host Microbe / Year: 2020Title: Human mAbs Broadly Protect against Arthritogenic Alphaviruses by Recognizing Conserved Elements of the Mxra8 Receptor-Binding Site. Authors: Laura A Powell / Andrew Miller / Julie M Fox / Nurgun Kose / Thomas Klose / Arthur S Kim / Robin Bombardi / Rashika N Tennekoon / A Dharshan de Silva / Robert H Carnahan / Michael S Diamond ...Authors: Laura A Powell / Andrew Miller / Julie M Fox / Nurgun Kose / Thomas Klose / Arthur S Kim / Robin Bombardi / Rashika N Tennekoon / A Dharshan de Silva / Robert H Carnahan / Michael S Diamond / Michael G Rossmann / Richard J Kuhn / James E Crowe /  Abstract: Mosquito inoculation of humans with arthritogenic alphaviruses results in a febrile syndrome characterized by debilitating musculoskeletal pain and arthritis. Despite an expanding global disease ...Mosquito inoculation of humans with arthritogenic alphaviruses results in a febrile syndrome characterized by debilitating musculoskeletal pain and arthritis. Despite an expanding global disease burden, no approved therapies or licensed vaccines exist. Here, we describe human monoclonal antibodies (mAbs) that bind to and neutralize multiple distantly related alphaviruses. These mAbs compete for an antigenic site and prevent attachment to the recently discovered Mxra8 alphavirus receptor. Three cryoelectron microscopy structures of Fab in complex with Ross River (RRV), Mayaro, or chikungunya viruses reveal a conserved footprint of the broadly neutralizing mAb RRV-12 in a region of the E2 glycoprotein B domain. This mAb neutralizes virus in vitro by preventing virus entry and spread and is protective in vivo in mouse models. Thus, the RRV-12 mAb and its defined epitope have potential as a therapeutic agent or target of vaccine design against multiple emerging arthritogenic alphavirus infections. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_21532.map.gz emd_21532.map.gz | 280.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-21532-v30.xml emd-21532-v30.xml emd-21532.xml emd-21532.xml | 17 KB 17 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_21532.png emd_21532.png | 255.3 KB | ||
| Filedesc metadata |  emd-21532.cif.gz emd-21532.cif.gz | 5.8 KB | ||
| Others |  emd_21532_half_map_1.map.gz emd_21532_half_map_1.map.gz emd_21532_half_map_2.map.gz emd_21532_half_map_2.map.gz | 125.8 MB 125.8 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-21532 http://ftp.pdbj.org/pub/emdb/structures/EMD-21532 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-21532 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-21532 | HTTPS FTP |
-Validation report
| Summary document |  emd_21532_validation.pdf.gz emd_21532_validation.pdf.gz | 909.7 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_21532_full_validation.pdf.gz emd_21532_full_validation.pdf.gz | 909.3 KB | Display | |
| Data in XML |  emd_21532_validation.xml.gz emd_21532_validation.xml.gz | 22.1 KB | Display | |
| Data in CIF |  emd_21532_validation.cif.gz emd_21532_validation.cif.gz | 26.7 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-21532 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-21532 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-21532 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-21532 | HTTPS FTP |
-Related structure data
| Related structure data |  6w2uMC  6vyvC  6w09C  6w1cC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_21532.map.gz / Format: CCP4 / Size: 1000 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_21532.map.gz / Format: CCP4 / Size: 1000 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | masked not sharpened | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.62 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
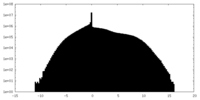
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Half map: half map not sharpened
| File | emd_21532_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map not sharpened | ||||||||||||
| Projections & Slices |
| ||||||||||||
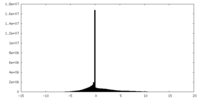
| Density Histograms |
-Half map: half map not sharpened
| File | emd_21532_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map not sharpened | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Native Mayaro Virus
| Entire | Name: Native Mayaro Virus |
|---|---|
| Components |
|
-Supramolecule #1: Native Mayaro Virus
| Supramolecule | Name: Native Mayaro Virus / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|
-Supramolecule #2: Mayaro virus
| Supramolecule | Name: Mayaro virus / type: complex / ID: 2 / Parent: 1 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Mayaro virus Mayaro virus |
-Macromolecule #1: Spike glycoprotein E1
| Macromolecule | Name: Spike glycoprotein E1 / type: protein_or_peptide / ID: 1 / Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Mayaro virus (strain Brazil) / Strain: Brazil Mayaro virus (strain Brazil) / Strain: Brazil |
| Molecular weight | Theoretical: 41.612867 KDa |
| Recombinant expression | Organism:  Mesocricetus auratus (golden hamster) Mesocricetus auratus (golden hamster) |
| Sequence | String: YEHTAIIPNQ VGFPYKAHVA REGYSPLTLQ MQVIETSLEP TLNLEYITCD YKTKVPSPYV KCCGTAECRT QDKPEYKCAV FTGVYPFMW GGAYCFCDSE NTQMSEAYVE RADVCKHDHA AAYRAHTASL RAKIKVTYGT VNQTVEAYVN GDHAVTIAGT K FIFGPVST ...String: YEHTAIIPNQ VGFPYKAHVA REGYSPLTLQ MQVIETSLEP TLNLEYITCD YKTKVPSPYV KCCGTAECRT QDKPEYKCAV FTGVYPFMW GGAYCFCDSE NTQMSEAYVE RADVCKHDHA AAYRAHTASL RAKIKVTYGT VNQTVEAYVN GDHAVTIAGT K FIFGPVST PWTPFDTKIL VYKGELYNQD FPRYGAGQPG RFGDIQSRTL DSRDLYANTG LKLARPAAGN IHVPYTQTPS GF KTWQKDR DSPLNAKAPF GCIIQTNPVR AMNCAVGNIP VSMDIADSAF TRLTDAPVIS ELTCTVSTCT HSSDFGGIAV LSY KVEKSG RCDIHSHSNV AVLQEVSIET EGRSVIHFST ASASPSFVVS VCSSRATCTA KCEP UniProtKB: Structural polyprotein |
-Macromolecule #2: Spike glycoprotein E2
| Macromolecule | Name: Spike glycoprotein E2 / type: protein_or_peptide / ID: 2 / Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Mayaro virus (strain Brazil) / Strain: Brazil Mayaro virus (strain Brazil) / Strain: Brazil |
| Molecular weight | Theoretical: 38.041957 KDa |
| Recombinant expression | Organism:  Mesocricetus auratus (golden hamster) Mesocricetus auratus (golden hamster) |
| Sequence | String: STANHFNAYK LTRPYVAYCA DCGMGHSCHS PAMIENIQAD ATDGTLKIQF ASQIGLTKTD THDHTKIRYA EGHDIAEAAR STLKVHSSS ECTVTGTMGH FILAKCPPGE RISVSFVDSK NEHRTCRIAY HHEQRLIGRE RFTVRPHHGI ELPCTTYQLT T AETSEEID ...String: STANHFNAYK LTRPYVAYCA DCGMGHSCHS PAMIENIQAD ATDGTLKIQF ASQIGLTKTD THDHTKIRYA EGHDIAEAAR STLKVHSSS ECTVTGTMGH FILAKCPPGE RISVSFVDSK NEHRTCRIAY HHEQRLIGRE RFTVRPHHGI ELPCTTYQLT T AETSEEID MHMPPDIPDR TILSQQSGNV KITVNGRTVR YSSSCGSQAV GTTTTDKTIN SCTVDKCQAY VTSHTKWQFN SP FVPRRMQ AERKGKVHIP FPLINTTCRV PLAPEALVRS GKREATLSLH PIHPTLLSYR TFGAERVFDE QWITAQTEVT IPV PVEGVE YQWGNHKPQR FVVA UniProtKB: Structural polyprotein |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 3 mg/mL |
|---|---|
| Buffer | pH: 7.5 |
| Grid | Model: C-flat / Support film - Material: CARBON / Support film - topology: HOLEY |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 85 % / Instrument: GATAN CRYOPLUNGE 3 |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 BASE (4k x 4k) / Average electron dose: 30.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 100.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal magnification: 18000 |
| Sample stage | Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: OTHER |
|---|---|
| Final reconstruction | Applied symmetry - Point group: I (icosahedral) / Resolution.type: BY AUTHOR / Resolution: 4.8 Å / Resolution method: FSC 0.143 CUT-OFF / Software - Name: jspr / Number images used: 18410 |
| Initial angle assignment | Type: RANDOM ASSIGNMENT |
| Final angle assignment | Type: PROJECTION MATCHING / Software - Name: jspr |
-Atomic model buiding 1
| Refinement | Protocol: RIGID BODY FIT |
|---|---|
| Output model |  PDB-6w2u: |
 Movie
Movie Controller
Controller



















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)