[English] 日本語
 Yorodumi
Yorodumi- PDB-1pge: PROSTAGLANDIN H2 SYNTHASE-1 COMPLEXED WITH P-(2'-IODO-5'-THENOYL)... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 1pge | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | PROSTAGLANDIN H2 SYNTHASE-1 COMPLEXED WITH P-(2'-IODO-5'-THENOYL)HYDROTROPIC ACID (IODOSUPROFEN) | |||||||||
 Components Components | PROSTAGLANDIN H2 SYNTHASE-1 | |||||||||
 Keywords Keywords | OXIDOREDUCTASE / DIOXYGENASE / PEROXIDASE | |||||||||
| Function / homology |  Function and homology information Function and homology informationprostaglandin-endoperoxide synthase / prostaglandin-endoperoxide synthase activity / cyclooxygenase pathway / oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen / prostaglandin biosynthetic process / peroxidase activity / regulation of blood pressure / response to oxidative stress / neuron projection / heme binding ...prostaglandin-endoperoxide synthase / prostaglandin-endoperoxide synthase activity / cyclooxygenase pathway / oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen / prostaglandin biosynthetic process / peroxidase activity / regulation of blood pressure / response to oxidative stress / neuron projection / heme binding / endoplasmic reticulum membrane / protein homodimerization activity / metal ion binding / cytoplasm Similarity search - Function | |||||||||
| Biological species |  | |||||||||
| Method |  X-RAY DIFFRACTION / Resolution: 3.5 Å X-RAY DIFFRACTION / Resolution: 3.5 Å | |||||||||
 Authors Authors | Loll, P.J. / Picot, D. / Garavito, R.M. | |||||||||
 Citation Citation |  Journal: Biochemistry / Year: 1996 Journal: Biochemistry / Year: 1996Title: Synthesis and use of iodinated nonsteroidal antiinflammatory drug analogs as crystallographic probes of the prostaglandin H2 synthase cyclooxygenase active site. Authors: Loll, P.J. / Picot, D. / Ekabo, O. / Garavito, R.M. #1:  Journal: Nat.Struct.Biol. / Year: 1995 Journal: Nat.Struct.Biol. / Year: 1995Title: The Structural Basis of Aspirin Activity Inferred from the Crystal Structure of Inactivated Prostaglandin H2 Synthase Authors: Loll, P.J. / Picot, D. / Garavito, R.M. #2:  Journal: Nature / Year: 1994 Journal: Nature / Year: 1994Title: The X-Ray Crystal Structure of the Membrane Protein Prostaglandin H2 Synthase-1 Authors: Picot, D. / Loll, P.J. / Garavito, R.M. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  1pge.cif.gz 1pge.cif.gz | 238 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb1pge.ent.gz pdb1pge.ent.gz | 191.5 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  1pge.json.gz 1pge.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Summary document |  1pge_validation.pdf.gz 1pge_validation.pdf.gz | 2.3 MB | Display |  wwPDB validaton report wwPDB validaton report |
|---|---|---|---|---|
| Full document |  1pge_full_validation.pdf.gz 1pge_full_validation.pdf.gz | 2.3 MB | Display | |
| Data in XML |  1pge_validation.xml.gz 1pge_validation.xml.gz | 49.1 KB | Display | |
| Data in CIF |  1pge_validation.cif.gz 1pge_validation.cif.gz | 62.6 KB | Display | |
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/pg/1pge https://data.pdbj.org/pub/pdb/validation_reports/pg/1pge ftp://data.pdbj.org/pub/pdb/validation_reports/pg/1pge ftp://data.pdbj.org/pub/pdb/validation_reports/pg/1pge | HTTPS FTP |
-Related structure data
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||
| Unit cell |
| ||||||||
| Noncrystallographic symmetry (NCS) | NCS oper: (Code: given Matrix: (-0.995479, -0.058108, 0.07513), Vector: |
- Components
Components
-Protein , 1 types, 2 molecules AB
| #1: Protein | Mass: 66164.812 Da / Num. of mol.: 2 / Source method: isolated from a natural source / Source: (natural)  References: PIR: A29947, UniProt: P05979*PLUS, prostaglandin-endoperoxide synthase |
|---|
-Sugars , 3 types, 8 molecules 


| #2: Polysaccharide | Source method: isolated from a genetically manipulated source #3: Sugar | ChemComp-NAG / #4: Sugar | |
|---|
-Non-polymers , 2 types, 4 molecules 
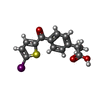

| #5: Chemical | | #6: Chemical | |
|---|
-Details
| Has protein modification | Y |
|---|---|
| Sequence details | THIS SEQUENCE IS FOUND IN ENTRY 1PRH. THE NUMBERING IS DERIVED FROM THE PRE-PROTEIN, WHICH CONTAINS ...THIS SEQUENCE IS FOUND IN ENTRY 1PRH. THE NUMBERING IS DERIVED FROM THE PRE-PROTEIN, WHICH CONTAINS A 24-RESIDUE SIGNAL SEQUENCE WHICH IS CLEAVED DURING MATURATION |
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION X-RAY DIFFRACTION |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 4.63 Å3/Da / Density % sol: 72 % | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Crystal grow | *PLUS Temperature: 18 ℃ / Method: vapor diffusion, hanging drop | ||||||||||||||||||||||||||||||||||||
| Components of the solutions | *PLUS
|
-Data collection
| Diffraction source | Wavelength: 1.5418 |
|---|---|
| Detector | Type: ENRAF-NONIUS FAST / Detector: DIFFRACTOMETER / Date: Apr 1, 1994 |
| Radiation | Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 1.5418 Å / Relative weight: 1 |
| Reflection | Resolution: 3.5→50 Å / Num. obs: 28057 / % possible obs: 89.8 % / Observed criterion σ(I): 0 / Redundancy: 2.5 % / Rmerge(I) obs: 0.104 |
| Reflection | *PLUS % possible obs: 90.9 % / Num. measured all: 69965 |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Resolution: 3.5→15 Å / σ(F): 1 Details: THE STARTING MODEL FOR THIS REFINEMENT WAS THE 3.1 ANGSTROM REFINED CRYSTAL STRUCTURE OF THE PROSTAGLANDIN SYNTHASE-FLURBIPROFEN COMPLEX, FOR WHICH INDIVIDUAL ISOTROPIC ATOMIC TEMPERATURE ...Details: THE STARTING MODEL FOR THIS REFINEMENT WAS THE 3.1 ANGSTROM REFINED CRYSTAL STRUCTURE OF THE PROSTAGLANDIN SYNTHASE-FLURBIPROFEN COMPLEX, FOR WHICH INDIVIDUAL ISOTROPIC ATOMIC TEMPERATURE FACTORS WERE REFINED. THESE B-VALUES WERE USED FOR THIS STRUCTURE WITHOUT FURTHER REFINEMENT.
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 22.2 Å2 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine analyze | Luzzati coordinate error obs: 0.3 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 3.5→15 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Software | *PLUS Name:  X-PLOR / Version: 3.1 / Classification: refinement X-PLOR / Version: 3.1 / Classification: refinement | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement | *PLUS Highest resolution: 3.5 Å / Lowest resolution: 8 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | *PLUS | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | *PLUS | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints | *PLUS
|
 Movie
Movie Controller
Controller























 PDBj
PDBj






