[English] 日本語
 Yorodumi
Yorodumi- PDB-1o2i: Elaborate Manifold of Short Hydrogen Bond Arrays Mediating Bindin... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 1o2i | ||||||
|---|---|---|---|---|---|---|---|
| Title | Elaborate Manifold of Short Hydrogen Bond Arrays Mediating Binding of Active Site-Directed Serine Protease Inhibitors | ||||||
 Components Components | BETA-TRYPSIN | ||||||
 Keywords Keywords | HYDROLASE / serine protease / short hydrogen bond / inhibition mechanism / shift of pKa / trypsin / thrombin / urokinase / factor Xa | ||||||
| Function / homology |  Function and homology information Function and homology informationtrypsin / serpin family protein binding / serine protease inhibitor complex / digestion / endopeptidase activity / serine-type endopeptidase activity / proteolysis / extracellular space / metal ion binding Similarity search - Function | ||||||
| Biological species |  | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  FOURIER SYNTHESIS / Resolution: 1.5 Å FOURIER SYNTHESIS / Resolution: 1.5 Å | ||||||
 Authors Authors | Katz, B.A. / Elrod, K. / Verner, E. / Mackman, R.L. / Luong, C. / Shrader, W. / Sendzik, M. / Spencer, J.R. / Sprengeler, P.A. / Kolesnikov, A. ...Katz, B.A. / Elrod, K. / Verner, E. / Mackman, R.L. / Luong, C. / Shrader, W. / Sendzik, M. / Spencer, J.R. / Sprengeler, P.A. / Kolesnikov, A. / Tai, W.F. / Hui, H. / Breitenbucher, G. / Allen, D. / Janc, J. | ||||||
 Citation Citation |  Journal: J.Mol.Biol. / Year: 2003 Journal: J.Mol.Biol. / Year: 2003Title: Elaborate Manifold of Short Hydrogen Bond Arrays Mediating Binding of Active Site-Directed Serine Protease Inhibitors Authors: Katz, B.A. / Elrod, K. / Verner, E. / Mackman, R.L. / Luong, C. / Shrader, W.D. / Sendzik, M. / Spencer, J.R. / Sprengeler, P.A. / Kolesnikov, A. / Tai, V.W.-F. / Hui, H.C. / Breitenbucher, ...Authors: Katz, B.A. / Elrod, K. / Verner, E. / Mackman, R.L. / Luong, C. / Shrader, W.D. / Sendzik, M. / Spencer, J.R. / Sprengeler, P.A. / Kolesnikov, A. / Tai, V.W.-F. / Hui, H.C. / Breitenbucher, J.G. / Allen, D. / Janc, J.W. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  1o2i.cif.gz 1o2i.cif.gz | 113.6 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb1o2i.ent.gz pdb1o2i.ent.gz | 89.5 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  1o2i.json.gz 1o2i.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Summary document |  1o2i_validation.pdf.gz 1o2i_validation.pdf.gz | 693.2 KB | Display |  wwPDB validaton report wwPDB validaton report |
|---|---|---|---|---|
| Full document |  1o2i_full_validation.pdf.gz 1o2i_full_validation.pdf.gz | 694.6 KB | Display | |
| Data in XML |  1o2i_validation.xml.gz 1o2i_validation.xml.gz | 14.4 KB | Display | |
| Data in CIF |  1o2i_validation.cif.gz 1o2i_validation.cif.gz | 21.7 KB | Display | |
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/o2/1o2i https://data.pdbj.org/pub/pdb/validation_reports/o2/1o2i ftp://data.pdbj.org/pub/pdb/validation_reports/o2/1o2i ftp://data.pdbj.org/pub/pdb/validation_reports/o2/1o2i | HTTPS FTP |
-Related structure data
| Related structure data |  1o2gC  1o2hC  1o2jC  1o2kC  1o2lC  1o2mC  1o2nC  1o2oC  1o2pC  1o2qC  1o2rC  1o2sC  1o2tC  1o2uC  1o2vC  1o2wC  1o2xC  1o2yC  1o2zC  1o30C  1o31C  1o32C  1o33C  1o34C  1o35C  1o36C  1o37C  1o38C  1o39C  1o3aC  1o3bC  1o3cC  1o3dC  1o3eC  1o3fC  1o3gC  1o3hC  1o3iC  1o3jC  1o3kC  1o3lC  1o3mC  1o3nC  1o3oC  1o3pC C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||||
| Unit cell |
|
- Components
Components
| #1: Protein | Mass: 23324.287 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  |
|---|---|
| #2: Chemical | ChemComp-CA / |
| #3: Chemical | ChemComp-SO4 / |
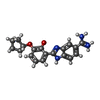
| #4: Chemical | ChemComp-655 / |
| #5: Water | ChemComp-HOH / |
| Has protein modification | Y |
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.99 Å3/Da / Density % sol: 58.9 % | |||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Crystal grow | Temperature: 298 K / Method: vapor diffusion / pH: 8.1 Details: magnesium sulfate soak at target pH (6.54). vapor diffusion at 298 K, pH 8.10 | |||||||||||||||||||||||||||||||||||
| Crystal grow | *PLUS Method: vapor diffusion, hanging drop / Details: Katz, B.A., (2001) J.Mol.Biol., 307, 1451. | |||||||||||||||||||||||||||||||||||
| Components of the solutions | *PLUS
|
-Data collection
| Diffraction | Mean temperature: 298 K |
|---|---|
| Diffraction source | Source:  ROTATING ANODE / Type: RIGAKU RU200 / Wavelength: 1.5418 ROTATING ANODE / Type: RIGAKU RU200 / Wavelength: 1.5418 |
| Detector | Type: RIGAKU RAXIS IV / Detector: IMAGE PLATE / Date: May 14, 2001 |
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 1.5418 Å / Relative weight: 1 |
| Reflection | Resolution: 1.5→28.92 Å / Num. all: 45626 / Num. obs: 34199 / % possible obs: 75 % / Observed criterion σ(I): 0.6 / Redundancy: 2 % / Rmerge(I) obs: 0.085 / Net I/σ(I): 8.8 |
| Reflection shell | Resolution: 1.5→1.57 Å / % possible obs: 43.3 % / Rmerge(I) obs: 0.299 / Mean I/σ(I) obs: 1.2 / Num. unique all: 5764 |
| Reflection | *PLUS Highest resolution: 1.4 Å / Num. obs: 35909 / Num. measured all: 70117 |
| Reflection shell | *PLUS Mean I/σ(I) obs: 1.3 |
- Processing
Processing
| Software |
| ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  FOURIER SYNTHESIS / Resolution: 1.5→7 Å / Cross valid method: THROUGHOUT / σ(F): 1.2 FOURIER SYNTHESIS / Resolution: 1.5→7 Å / Cross valid method: THROUGHOUT / σ(F): 1.2 Details: Residues simultaneously refined in two or more conformations are: Val53, Leu66, Ser86, Ser110, Ser113, Ser130, Lys159, Asp165, Lys169, Ser170, Gln175, Lys230, Ser236, Ser244 Note that HOH383 ...Details: Residues simultaneously refined in two or more conformations are: Val53, Leu66, Ser86, Ser110, Ser113, Ser130, Lys159, Asp165, Lys169, Ser170, Gln175, Lys230, Ser236, Ser244 Note that HOH383 makes short H-bonds to OgSer195 and O6' of the inhibitor Disordered waters are: HOH276 which is close to conformation #2 of Lys230; HOH301 which is close to conformation #1 of Asp165 His40 and HIS91 are MONOPROTONATED ON THE EPSILON NITROGEN. His57 is doubly protonated.
| ||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 1.5→7 Å
| ||||||||||||||||||||
| Refine LS restraints |
| ||||||||||||||||||||
| Refinement | *PLUS Highest resolution: 1.5 Å / % reflection Rfree: 10 % / Rfactor Rwork: 0.192 | ||||||||||||||||||||
| Solvent computation | *PLUS | ||||||||||||||||||||
| Displacement parameters | *PLUS | ||||||||||||||||||||
| Refine LS restraints | *PLUS
| ||||||||||||||||||||
| LS refinement shell | *PLUS Highest resolution: 1.5 Å / Lowest resolution: 1.57 Å / Rfactor Rfree: 0.211 |
 Movie
Movie Controller
Controller






















 PDBj
PDBj