[English] 日本語
 Yorodumi
Yorodumi- PDB-1bph: CONFORMATIONAL CHANGES IN CUBIC INSULIN CRYSTALS IN THE PH RANGE 7-11 -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 1bph | ||||||
|---|---|---|---|---|---|---|---|
| Title | CONFORMATIONAL CHANGES IN CUBIC INSULIN CRYSTALS IN THE PH RANGE 7-11 | ||||||
 Components Components |
| ||||||
 Keywords Keywords | HORMONE | ||||||
| Function / homology |  Function and homology information Function and homology informationestradiol secretion / positive regulation of blood circulation / negative regulation of lactation / glucose import in response to insulin stimulus / positive regulation of cell maturation / positive regulation of lactation / response to L-arginine / positive regulation of mammary gland epithelial cell proliferation / negative regulation of appetite / response to butyrate ...estradiol secretion / positive regulation of blood circulation / negative regulation of lactation / glucose import in response to insulin stimulus / positive regulation of cell maturation / positive regulation of lactation / response to L-arginine / positive regulation of mammary gland epithelial cell proliferation / negative regulation of appetite / response to butyrate / feeding behavior / response to growth hormone / response to food / positive regulation of peptide hormone secretion / positive regulation of Rho protein signal transduction / protein secretion / negative regulation of lipid catabolic process / response to glucose / positive regulation of protein secretion / insulin receptor binding / response to nutrient levels / hormone activity / positive regulation of insulin secretion / glucose metabolic process / glucose homeostasis / response to heat / positive regulation of phosphatidylinositol 3-kinase/protein kinase B signal transduction / positive regulation of gene expression / negative regulation of apoptotic process / extracellular space / identical protein binding Similarity search - Function | ||||||
| Biological species |  | ||||||
| Method |  X-RAY DIFFRACTION / Resolution: 2 Å X-RAY DIFFRACTION / Resolution: 2 Å | ||||||
 Authors Authors | Gursky, O. / Badger, J. / Li, Y. / Caspar, D.L.D. | ||||||
 Citation Citation |  Journal: Biophys.J. / Year: 1992 Journal: Biophys.J. / Year: 1992Title: Conformational changes in cubic insulin crystals in the pH range 7-11. Authors: Gursky, O. / Badger, J. / Li, Y. / Caspar, D.L. #1:  Journal: Biophys.J. / Year: 1992 Journal: Biophys.J. / Year: 1992Title: Monovalent Cation Binding in Cubic Insulin Crystals Authors: Gursky, O. / Li, Y. / Badger, J. / Caspar, D.L.D. #3:  Journal: Acta Crystallogr.,Sect.B / Year: 1991 Journal: Acta Crystallogr.,Sect.B / Year: 1991Title: Structure of the Pig Insulin Dimer in the Cubic Crystal Authors: Badger, J. / Harris, M.R. / Reynolds, C.D. / Evans, A.C. / Dodson, E.J. / Dodson, G.G. / North, A.C.T. #4:  Journal: Proc.Natl.Acad.Sci.USA / Year: 1991 Journal: Proc.Natl.Acad.Sci.USA / Year: 1991Title: Water Structure in Cubic Insulin Crystals Authors: Badger, J. / Caspar, D.L.D. #5:  Journal: J.Mol.Biol. / Year: 1978 Journal: J.Mol.Biol. / Year: 1978Title: Zinc-Free Cubic Pig Insulin: Crystallization and Structure Determination Authors: Dodson, E.J. / Dodson, G.G. / Lewitova, A. / Sabesan, M. | ||||||
| History |
| ||||||
| Remark 700 | SHEET THERE IS A SHEET COMPRISING TWO ANTIPARALLEL STRANDS PHE B 24 - TYR B 26 FROM TWO DIMER- ...SHEET THERE IS A SHEET COMPRISING TWO ANTIPARALLEL STRANDS PHE B 24 - TYR B 26 FROM TWO DIMER-FORMING INSULIN MOLECULES. |
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  1bph.cif.gz 1bph.cif.gz | 25.4 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb1bph.ent.gz pdb1bph.ent.gz | 16.6 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  1bph.json.gz 1bph.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/bp/1bph https://data.pdbj.org/pub/pdb/validation_reports/bp/1bph ftp://data.pdbj.org/pub/pdb/validation_reports/bp/1bph ftp://data.pdbj.org/pub/pdb/validation_reports/bp/1bph | HTTPS FTP |
|---|
-Related structure data
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||
| 2 | 
| ||||||||
| 3 | 
| ||||||||
| 4 | 
| ||||||||
| Unit cell |
| ||||||||
| Components on special symmetry positions |
|
- Components
Components
| #1: Protein/peptide | Mass: 2339.645 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  |
|---|---|
| #2: Protein/peptide | Mass: 3403.927 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  |
| #3: Chemical | ChemComp-NA / |
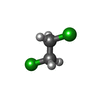
| #4: Chemical | ChemComp-DCE / |
| #5: Water | ChemComp-HOH / |
| Has protein modification | Y |
| Nonpolymer details | THE 1,2-DICHLOROETHANE IS BOUND IS CIS CONFORMATION IN A SYMMETRIC POSITION ACROSS THE ...THE 1,2-DICHLOROET |
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION X-RAY DIFFRACTION |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 3.56 Å3/Da / Density % sol: 65.47 % | ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Crystal grow | *PLUS pH: 9.2 / Method: microdialysisDetails: referred to 'Dodson, E.J.', (1978) J.Mol.Biol., 125, 387-396 | ||||||||||||||||||||||||
| Components of the solutions | *PLUS
|
-Data collection
| Radiation | Scattering type: x-ray |
|---|---|
| Radiation wavelength | Relative weight: 1 |
| Reflection | *PLUS Highest resolution: 1.9 Å / Num. obs: 5760 / % possible obs: 93 % / Num. measured all: 21890 / Rmerge(I) obs: 0.053 |
- Processing
Processing
| Software | Name: PROLSQ / Classification: refinement | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Rfactor obs: 0.16 / Highest resolution: 2 Å Details: IN 1BPH AND 1CPH, THE SIDE CHAIN OF GLU A 4 CAN ADOPT TWO ALTERNATIVE POSITIONS WHICH OVERLAP. THEIR RELATIVE WEIGHT AND THE ATOMIC POSITIONS OF THE SECOND CONFORMER ARE NOT ACCURATELY ...Details: IN 1BPH AND 1CPH, THE SIDE CHAIN OF GLU A 4 CAN ADOPT TWO ALTERNATIVE POSITIONS WHICH OVERLAP. THEIR RELATIVE WEIGHT AND THE ATOMIC POSITIONS OF THE SECOND CONFORMER ARE NOT ACCURATELY DETERMINED. IN 1APH, 1BPH, AND 1DPH, THE SIDE CHAIN OF GLU B 21 IS DISORDERED. IT HAS BEEN MODELED AS SUPERPOSITION OF TWO CONFORMATIONS BUT ATOMIC POSITIONS FOR THESE CONFORMATIONS ARE PROBABLY NOT VERY ACCURATE. THE SIDE CHAIN OF LYS B 29 IS POORLY DEFINED IN THE ELECTRON DENSITY MAPS. IN 1APH AND 1CPH, IT IS INCLUDED WITH PARTIAL OCCUPANCY. IN 1BPH AND 1DPH, ITS COORDINATES HAVE BEEN OMITTED FROM THE ENTRY. THE MAIN AND SIDE CHAIN OF ALA B 30 (C-TERMINAL RESIDUE OF CHAIN B) CAN ADOPT TWO SEPARATE CONFORMATIONS AND IS DISORDERED IN EACH OF THESE CONFORMATIONS, WHICH LIMITED THE ACCURACY OF DETERMINATION OF ATOMIC POSITIONS FOR THE CONFORMERS OF ALA B 30. IN 1APH AND 1CPH, SINGLE ALTERNATIVE CONFORMERS ARE PREDOMINANT BUT, DUE TO DISORDER, THEY ARE ASSIGNED PARTIAL OCCUPANCIES. IN 1BPH AND 1DPH, BOTH ALTERNATIVE CONFORMERS ARE INCLUDED IN THE ENTRY. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Highest resolution: 2 Å
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement | *PLUS Highest resolution: 2 Å / Rfactor obs: 0.16 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | *PLUS | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | *PLUS | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints | *PLUS
|
 Movie
Movie Controller
Controller















 PDBj
PDBj