+ データを開く
データを開く
- 基本情報
基本情報
| 登録情報 | データベース: PDB / ID: 193d | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
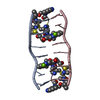
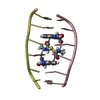
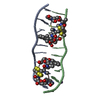
| タイトル | SOLUTION STRUCTURE OF A QUINOMYCIN BISINTERCALATOR-DNA COMPLEX | |||||||||
 要素 要素 |
| |||||||||
 キーワード キーワード | DNA/ANTIBIOTIC / BISINTERCALATOR / DEPSIPEPTIDE / QUINOXALINE / THIOACETAL / ANTIBIOTIC / ANTITUMOR / DNA-ANTIBIOTIC COMPLEX | |||||||||
| 機能・相同性 | uk63052 / 3-HYDROXYQUINALDIC ACID / DNA 機能・相同性情報 機能・相同性情報 | |||||||||
| 生物種 |  STREPTOMYCES (バクテリア) STREPTOMYCES (バクテリア) | |||||||||
| 手法 | 溶液NMR / MOLECULAR DYNAMICS, MATRIX RELAXATION | |||||||||
 データ登録者 データ登録者 | Chen, H. / Patel, D.J. | |||||||||
 引用 引用 |  ジャーナル: J.Mol.Biol. / 年: 1995 ジャーナル: J.Mol.Biol. / 年: 1995タイトル: Solution Structure of a Quinomycin Bisintercalator-DNA Complex. 著者: Chen, H. / Patel, D.J. | |||||||||
| 履歴 |
|
- 構造の表示
構造の表示
| 構造ビューア | 分子:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- ダウンロードとリンク
ダウンロードとリンク
- ダウンロード
ダウンロード
| PDBx/mmCIF形式 |  193d.cif.gz 193d.cif.gz | 64.9 KB | 表示 |  PDBx/mmCIF形式 PDBx/mmCIF形式 |
|---|---|---|---|---|
| PDB形式 |  pdb193d.ent.gz pdb193d.ent.gz | 53.7 KB | 表示 |  PDB形式 PDB形式 |
| PDBx/mmJSON形式 |  193d.json.gz 193d.json.gz | ツリー表示 |  PDBx/mmJSON形式 PDBx/mmJSON形式 | |
| その他 |  その他のダウンロード その他のダウンロード |
-検証レポート
| 文書・要旨 |  193d_validation.pdf.gz 193d_validation.pdf.gz | 377 KB | 表示 |  wwPDB検証レポート wwPDB検証レポート |
|---|---|---|---|---|
| 文書・詳細版 |  193d_full_validation.pdf.gz 193d_full_validation.pdf.gz | 419.1 KB | 表示 | |
| XML形式データ |  193d_validation.xml.gz 193d_validation.xml.gz | 6.8 KB | 表示 | |
| CIF形式データ |  193d_validation.cif.gz 193d_validation.cif.gz | 8.6 KB | 表示 | |
| アーカイブディレクトリ |  https://data.pdbj.org/pub/pdb/validation_reports/93/193d https://data.pdbj.org/pub/pdb/validation_reports/93/193d ftp://data.pdbj.org/pub/pdb/validation_reports/93/193d ftp://data.pdbj.org/pub/pdb/validation_reports/93/193d | HTTPS FTP |
-関連構造データ
| 関連構造データ | |
|---|---|
| 類似構造データ |
- リンク
リンク
- 集合体
集合体
| 登録構造単位 | 
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| |||||||||
| Atom site foot note | 1: ALA D 2 - CYS D 3 MODEL 1 OMEGA = 0.00 PEPTIDE BOND DEVIATES SIGNIFICANTLY FROM TRANS CONFORMATION 2: ALA C 2 - CYS C 3 MODEL 2 OMEGA = 0.00 PEPTIDE BOND DEVIATES SIGNIFICANTLY FROM TRANS CONFORMATION 3: ALA D 2 - CYS D 3 MODEL 3 OMEGA = 0.00 PEPTIDE BOND DEVIATES SIGNIFICANTLY FROM TRANS CONFORMATION 4: ALA D 2 - CYS D 3 MODEL 4 OMEGA = 359.98 PEPTIDE BOND DEVIATES SIGNIFICANTLY FROM TRANS CONFORMATION 7: SER C 1 IS A D-SERINE. / 8: SER D 1 IS A D-SERINE. | |||||||||
| NMR アンサンブル |
|
- 要素
要素
| #1: DNA鎖 | 分子量: 2426.617 Da / 分子数: 2 / 由来タイプ: 合成 / 詳細: CHEMICALLY SYNTHESIZED #2: タンパク質・ペプチド | | #3: 化合物 | 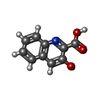  タイプ: Cyclic depsipeptide / クラス: 抗生剤 / 分子量: 189.167 Da / 分子数: 2 / 由来タイプ: 合成 / 式: C10H7NO3 タイプ: Cyclic depsipeptide / クラス: 抗生剤 / 分子量: 189.167 Da / 分子数: 2 / 由来タイプ: 合成 / 式: C10H7NO3詳細: QUINOMYCIN UK63052 IS A BICYCLIC OCTADEPSIPEPTIDE. BICYCLIZATION IS ACHIEVED BY LINKING THE N- AND THE C- TERMINI, AND A THIOACETAL BOND BETWEEN RESIDUES 3 AND 7. THE TWO HYDROXYQUINOXALINE ...詳細: QUINOMYCIN UK63052 IS A BICYCLIC OCTADEPSIPEPTIDE. BICYCLIZATION IS ACHIEVED BY LINKING THE N- AND THE C- TERMINI, AND A THIOACETAL BOND BETWEEN RESIDUES 3 AND 7. THE TWO HYDROXYQUINOXALINE CHROMOPHORES ARE LINKED TO THE D-SERINE RESIDUES, RESIDUES 1 AND 5. 参照: uk63052 構成要素の詳細 | QUINOMYCIN UK63052 IS A BICYCLIC OCTADEPSIPEPTIDE, A MEMBER OF THE QUINOXALINE CLASS OF ANTIBIOTICS. ...QUINOMYCIN | |
|---|
-実験情報
-実験
| 実験 | 手法: 溶液NMR |
|---|
- 試料調製
試料調製
| 詳細 | 溶媒系: D2O |
|---|---|
| 結晶化 | *PLUS 手法: other / 詳細: NMR |
- 解析
解析
| ソフトウェア |
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| NMR software | 名称:  X-PLOR / 開発者: BRUNGER / 分類: 精密化 X-PLOR / 開発者: BRUNGER / 分類: 精密化 | ||||||||
| 精密化 | 手法: MOLECULAR DYNAMICS, MATRIX RELAXATION / ソフトェア番号: 1 詳細: TWO STARTING STRUCTURES WERE GENERATED BY MANUALLY DOCKING THE DRUG ONTO A- AND B-FORM DNA USING INSIGHT II AND WERE SUBSEQUENTLY REFINED BY DISTANCE- RESTRAINED MOLECULAR DYNAMICS USING A ...詳細: TWO STARTING STRUCTURES WERE GENERATED BY MANUALLY DOCKING THE DRUG ONTO A- AND B-FORM DNA USING INSIGHT II AND WERE SUBSEQUENTLY REFINED BY DISTANCE- RESTRAINED MOLECULAR DYNAMICS USING A SET OF INTER-PROTON DISTANCE RESTRAINTS DERIVED FROM THE NMR DATA. TWO INITIAL VELOCITY SEEDS WERE USED FOR EACH STARTING STRUCTURE WHICH YIELDS FOUR DISTANCE-REFINED STRUCTURES. THEY WERE REFINED FURTHER USING RELAXATION-MATRIX BASED NOE INTENSITY-RESTRAINED MOLECULAR DYNAMICS. THE FINAL FOUR STRUCTURES WERE OBTAINED BY TAKING THE AVERAGE COORDINATES OF THE LAST 0.5 PS OF THE DYNAMICS DURING RELAXATION MATRIX REFINEMENT AND ENERGY MINIMIZED. THE R(1/6) VALUE WAS USED TO REFINE THE STRUCTURE DURING RELAXATION MATRIX REFINEMENT. THE SUMMATIONS RUN THROUGH ALL OBSERVED, QUANTIFIABLE NOE CROSSPEAKS IN NOESY SPECTRA RECORDED IN D2O AND MIXING TIMES OF 30, 60, 120 AND 180 MS. THE R(1/6) FACTOR AND THE RMS DEVIATIONS FROM IDEAL GEOMETRY FOR THE FOUR FINAL STRUCTURES ARE: MODEL1 MODEL2 MODEL3 MODEL4 R(1/6) FACTOR 0.023 0.024 0.024 0.026 BOND (ANG) 0.011 0.011 0.011 0.011 ANGLES (DEG) 3.616 3.657 3.700 3.725 IMPROPERS (DEG) 0.274 0.285 0.241 0.298 | ||||||||
| NMRアンサンブル | コンフォーマー選択の基準: all calculated structures submitted 計算したコンフォーマーの数: 4 / 登録したコンフォーマーの数: 4 |
 ムービー
ムービー コントローラー
コントローラー













 PDBj
PDBj

