[English] 日本語
 Yorodumi
Yorodumi- EMDB-1730: 4.6 Angstrom Cryo-EM reconstruction of Tobacco Mosaic Virus from ... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-1730 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | 4.6 Angstrom Cryo-EM reconstruction of Tobacco Mosaic Virus from images recorded at 300 KeV on a 4kx4k CCD camera | |||||||||
 Map data Map data | B-factor scaled fully CTF corrected density map of TMV | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Cryo-EM / single particle processing / TMV / CCD data / 300 KeV electrons | |||||||||
| Function / homology | Tobacco mosaic virus-like, coat protein / Tobacco mosaic virus-like, coat protein superfamily / Virus coat protein (TMV like) / helical viral capsid / structural molecule activity / Capsid protein Function and homology information Function and homology information | |||||||||
| Biological species |   Tobacco mosaic virus Tobacco mosaic virus | |||||||||
| Method | helical reconstruction / cryo EM / negative staining / Resolution: 4.6 Å | |||||||||
 Authors Authors | Clare DK / Orlova EV | |||||||||
 Citation Citation |  Journal: J Struct Biol / Year: 2010 Journal: J Struct Biol / Year: 2010Title: 4.6A Cryo-EM reconstruction of tobacco mosaic virus from images recorded at 300 keV on a 4k x 4k CCD camera. Authors: Daniel K Clare / Elena V Orlova /  Abstract: Tobacco mosaic virus (TMV) is a plant virus with a highly ordered organisation and has been described in three different structural states: As stacked disks without RNA (X-ray crystallography), as a ...Tobacco mosaic virus (TMV) is a plant virus with a highly ordered organisation and has been described in three different structural states: As stacked disks without RNA (X-ray crystallography), as a helical form with RNA (X-ray fibre diffraction) and as a second distinct helical form with RNA (cryo-EM). Here we present a structural analysis of TMV as a test object to assess the quality of cryo-EM images recorded at 300 keV on a CCD camera. The 4.6A TMV structure obtained is consistent with the previous cryo-EM structure and confirms that there is a second helical form of TMV. The structure here also shows that with a similar number of TMV segments an equivalent resolution can be achieved with a 4k CCD camera at 300 keV. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_1730.map.gz emd_1730.map.gz | 16.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-1730-v30.xml emd-1730-v30.xml emd-1730.xml emd-1730.xml | 12.2 KB 12.2 KB | Display Display |  EMDB header EMDB header |
| Images |  1730.png 1730.png | 492.2 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-1730 http://ftp.pdbj.org/pub/emdb/structures/EMD-1730 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-1730 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-1730 | HTTPS FTP |
-Related structure data
| Related structure data |  2xeaMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_1730.map.gz / Format: CCP4 / Size: 29.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_1730.map.gz / Format: CCP4 / Size: 29.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | B-factor scaled fully CTF corrected density map of TMV | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.24 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
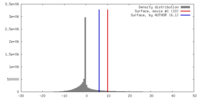
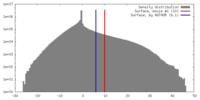
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Tobacco Mosaic Virus
| Entire | Name:   Tobacco Mosaic Virus Tobacco Mosaic Virus |
|---|---|
| Components |
|
-Supramolecule #1000: Tobacco Mosaic Virus
| Supramolecule | Name: Tobacco Mosaic Virus / type: sample / ID: 1000 / Details: Monodisperse Oligomeric state: Homo-oligomer of TMV coat protein and ssRNA Number unique components: 2 |
|---|---|
| Molecular weight | Experimental: 175 KDa / Theoretical: 175 KDa |
-Supramolecule #1: Tobacco mosaic virus
| Supramolecule | Name: Tobacco mosaic virus / type: virus / ID: 1 / Name.synonym: TMV / NCBI-ID: 12242 / Sci species name: Tobacco mosaic virus / Database: NCBI / Virus type: VIRION / Virus isolate: OTHER / Virus enveloped: No / Virus empty: No / Syn species name: TMV |
|---|---|
| Host (natural) | Organism:  |
| Molecular weight | Experimental: 175 KDa / Theoretical: 175 KDa |
| Virus shell | Shell ID: 1 / Name: CP / Diameter: 180 Å |
-Macromolecule #1: ssRNA
| Macromolecule | Name: ssRNA / type: rna / ID: 1 / Name.synonym: RNA / Classification: OTHER / Structure: SINGLE STRANDED / Synthetic?: No |
|---|---|
| Source (natural) | Organism:   Tobacco mosaic virus / synonym: TMV Tobacco mosaic virus / synonym: TMV |
-Experimental details
-Structure determination
| Method | negative staining, cryo EM |
|---|---|
 Processing Processing | helical reconstruction |
| Aggregation state | filament |
- Sample preparation
Sample preparation
| Concentration | 3 mg/mL |
|---|---|
| Buffer | pH: 7.4 / Details: 50 mM Tris-HCl, 50 mM KCl, 10 mM MgCl2 |
| Staining | Type: NEGATIVE Details: 3.5ul of TMV was added to R2/2 C-flat grid, blotted, then plunge frozen in liquid ethane |
| Grid | Details: 400 mesh R2/2 c-flat grids |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 60 % / Chamber temperature: 100 K / Instrument: HOMEMADE PLUNGER / Details: Vitrification instrument: Home made plunger Method: Grids were blotted for around 2 seconds and then rapidly plunged into liquid ethane |
- Electron microscopy
Electron microscopy
| Microscope | FEI POLARA 300 |
|---|---|
| Temperature | Average: 78 K |
| Alignment procedure | Legacy - Astigmatism: Corrected at 115,000 times |
| Date | Feb 1, 2009 |
| Image recording | Category: CCD / Film or detector model: GATAN ULTRASCAN 4000 (4k x 4k) / Digitization - Sampling interval: 15 µm / Number real images: 104 / Average electron dose: 25 e/Å2 / Bits/pixel: 16 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated magnification: 121000 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.3 mm / Nominal defocus max: 3.0 µm / Nominal defocus min: 0.9 µm / Nominal magnification: 90000 |
| Sample stage | Specimen holder: Eucentric / Specimen holder model: GATAN HELIUM |
| Experimental equipment |  Model: Tecnai Polara / Image courtesy: FEI Company |
- Image processing
Image processing
| Details | The particle were selected using the helical option in boxer |
|---|---|
| Final reconstruction | Applied symmetry - Helical parameters - Δz: 1.408 Å Applied symmetry - Helical parameters - Δ&Phi: 22.04 ° Applied symmetry - Helical parameters - Axial symmetry: C1 (asymmetric) Algorithm: OTHER / Resolution.type: BY AUTHOR / Resolution: 4.6 Å / Resolution method: FSC 0.5 CUT-OFF / Software - Name: SPIDER, IMAGIC / Details: BPRP was used for reconstruction |
| CTF correction | Details: Each particle was fully CTF corrected |
| Final angle assignment | Details: 80-100 in theta and 0-22.04 in phi in 0.5 steps |
-Atomic model buiding 1
| Initial model | PDB ID: |
|---|---|
| Software | Name: Chimera and Coot |
| Details | Protocol: Rigid body, real space. The PDB was initially fitted using chimera, the coordinates were then refined using Coot |
| Refinement | Space: REAL / Protocol: RIGID BODY FIT / Target criteria: Cross-correlation coefficient |
| Output model |  PDB-2xea: |
 Movie
Movie Controller
Controller









 Z (Sec.)
Z (Sec.) X (Row.)
X (Row.) Y (Col.)
Y (Col.)