[English] 日本語
 Yorodumi
Yorodumi- EMDB-16367: General transcription factor IIH in open conformation focused map -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
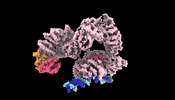
| Title | General transcription factor IIH in open conformation focused map | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Mammalian PIC / +1 nucleosome / transcription initiation / TRANSCRIPTION | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
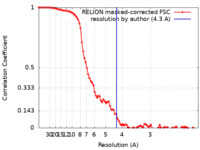
| Method | single particle reconstruction / cryo EM / Resolution: 4.3 Å | |||||||||
 Authors Authors | Abril-Garrido J / Dienemann C / Grabbe F / Velychko T / Lidschreiber M / Wang H / Cramer P | |||||||||
| Funding support |  Germany, European Union, 2 items Germany, European Union, 2 items
| |||||||||
 Citation Citation |  Journal: Mol Cell / Year: 2023 Journal: Mol Cell / Year: 2023Title: Structural basis of transcription reduction by a promoter-proximal +1 nucleosome. Authors: Julio Abril-Garrido / Christian Dienemann / Frauke Grabbe / Taras Velychko / Michael Lidschreiber / Haibo Wang / Patrick Cramer /  Abstract: At active human genes, the +1 nucleosome is located downstream of the RNA polymerase II (RNA Pol II) pre-initiation complex (PIC). However, at inactive genes, the +1 nucleosome is found further ...At active human genes, the +1 nucleosome is located downstream of the RNA polymerase II (RNA Pol II) pre-initiation complex (PIC). However, at inactive genes, the +1 nucleosome is found further upstream, at a promoter-proximal location. Here, we establish a model system to show that a promoter-proximal +1 nucleosome can reduce RNA synthesis in vivo and in vitro, and we analyze its structural basis. We find that the PIC assembles normally when the edge of the +1 nucleosome is located 18 base pairs (bp) downstream of the transcription start site (TSS). However, when the nucleosome edge is located further upstream, only 10 bp downstream of the TSS, the PIC adopts an inhibited state. The transcription factor IIH (TFIIH) shows a closed conformation and its subunit XPB contacts DNA with only one of its two ATPase lobes, inconsistent with DNA opening. These results provide a mechanism for nucleosome-dependent regulation of transcription initiation. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_16367.map.gz emd_16367.map.gz | 46 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-16367-v30.xml emd-16367-v30.xml emd-16367.xml emd-16367.xml | 34.2 KB 34.2 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_16367_fsc.xml emd_16367_fsc.xml | 8.9 KB | Display |  FSC data file FSC data file |
| Images |  emd_16367.png emd_16367.png | 56.4 KB | ||
| Masks |  emd_16367_msk_1.map emd_16367_msk_1.map emd_16367_msk_2.map emd_16367_msk_2.map emd_16367_msk_3.map emd_16367_msk_3.map | 59.6 MB 59.6 MB 59.6 MB |  Mask map Mask map | |
| Others |  emd_16367_additional_1.map.gz emd_16367_additional_1.map.gz emd_16367_additional_2.map.gz emd_16367_additional_2.map.gz emd_16367_additional_3.map.gz emd_16367_additional_3.map.gz emd_16367_additional_4.map.gz emd_16367_additional_4.map.gz emd_16367_additional_5.map.gz emd_16367_additional_5.map.gz emd_16367_additional_6.map.gz emd_16367_additional_6.map.gz emd_16367_additional_7.map.gz emd_16367_additional_7.map.gz emd_16367_additional_8.map.gz emd_16367_additional_8.map.gz emd_16367_additional_9.map.gz emd_16367_additional_9.map.gz emd_16367_half_map_1.map.gz emd_16367_half_map_1.map.gz emd_16367_half_map_2.map.gz emd_16367_half_map_2.map.gz | 46.2 MB 55.7 MB 45.8 MB 55.6 MB 46.2 MB 46.2 MB 45.9 MB 55.8 MB 46.2 MB 46.2 MB 46.2 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-16367 http://ftp.pdbj.org/pub/emdb/structures/EMD-16367 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-16367 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-16367 | HTTPS FTP |
-Validation report
| Summary document |  emd_16367_validation.pdf.gz emd_16367_validation.pdf.gz | 849.6 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_16367_full_validation.pdf.gz emd_16367_full_validation.pdf.gz | 849.1 KB | Display | |
| Data in XML |  emd_16367_validation.xml.gz emd_16367_validation.xml.gz | 14.3 KB | Display | |
| Data in CIF |  emd_16367_validation.cif.gz emd_16367_validation.cif.gz | 20.3 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-16367 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-16367 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-16367 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-16367 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_16367.map.gz / Format: CCP4 / Size: 59.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_16367.map.gz / Format: CCP4 / Size: 59.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.05 Å | ||||||||||||||||||||||||||||||||||||
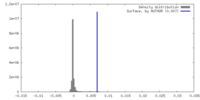
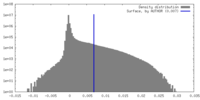
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
+Mask #1
+Mask #2
+Mask #3
+Additional map: #1
+Additional map: #2
+Additional map: #3
+Additional map: #4
+Additional map: #5
+Additional map: #6
+Additional map: #7
+Additional map: #8
+Additional map: #9
+Half map: #1
+Half map: #2
- Sample components
Sample components
-Entire : General transcription factor IIH
| Entire | Name: General transcription factor IIH |
|---|---|
| Components |
|
-Supramolecule #1: General transcription factor IIH
| Supramolecule | Name: General transcription factor IIH / type: complex / ID: 1 / Parent: 0 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 370 KDa |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Grid | Model: Quantifoil R3.5/1 / Material: COPPER / Support film - Material: CARBON / Support film - topology: CONTINUOUS / Support film - Film thickness: 3 |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Quantum SE / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Number real images: 41517 / Average exposure time: 3.0 sec. / Average electron dose: 41.58 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 1.5 µm / Nominal defocus min: 0.5 µm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller




















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)