Entry Database : EMDB / ID : EMD-16331Title RNA polymerase II pre-initiation complex with the proximal +1 nucleosome (PIC-Nuc10W) Complex : RNA polymerase II pre-initiation complex with the proximal +1 nucleosomeComplex : General transcription factor IIHProtein or peptide : x 6 typesComplex : Mammalian RNA polymerase IIProtein or peptide : x 6 typesComplex : Nucleosome (protein)Protein or peptide : x 4 typesComplex : Nucleosome (DNA)Complex : General transcription factor IIBProtein or peptide : x 1 typesComplex : TATA-box binding proteinProtein or peptide : x 1 typesComplex : General transcription factor IIFProtein or peptide : x 2 typesComplex : General transcription factor IIAProtein or peptide : x 2 typesComplex : General transcription factor IIEProtein or peptide : x 1 typesProtein or peptide : x 11 typesLigand : x 3 types / / / Function / homology Function Domain/homology Component
/ / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / Biological species Homo sapiens (human) / Sus scrofa (pig) / Xenopus laevis (African clawed frog) / Method / / Resolution : 4.1 Å Abril-Garrido J / Dienemann C / Grabbe F / Velychko T / Lidschreiber M / Wang H / Cramer P Funding support Organization Grant number Country German Research Foundation (DFG) European Research Council (ERC) European Union
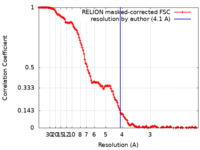
Journal : Mol Cell / Year : 2023Title : Structural basis of transcription reduction by a promoter-proximal +1 nucleosome.Authors : Julio Abril-Garrido / Christian Dienemann / Frauke Grabbe / Taras Velychko / Michael Lidschreiber / Haibo Wang / Patrick Cramer / Abstract : At active human genes, the +1 nucleosome is located downstream of the RNA polymerase II (RNA Pol II) pre-initiation complex (PIC). However, at inactive genes, the +1 nucleosome is found further ... At active human genes, the +1 nucleosome is located downstream of the RNA polymerase II (RNA Pol II) pre-initiation complex (PIC). However, at inactive genes, the +1 nucleosome is found further upstream, at a promoter-proximal location. Here, we establish a model system to show that a promoter-proximal +1 nucleosome can reduce RNA synthesis in vivo and in vitro, and we analyze its structural basis. We find that the PIC assembles normally when the edge of the +1 nucleosome is located 18 base pairs (bp) downstream of the transcription start site (TSS). However, when the nucleosome edge is located further upstream, only 10 bp downstream of the TSS, the PIC adopts an inhibited state. The transcription factor IIH (TFIIH) shows a closed conformation and its subunit XPB contacts DNA with only one of its two ATPase lobes, inconsistent with DNA opening. These results provide a mechanism for nucleosome-dependent regulation of transcription initiation. History Deposition Dec 14, 2022 - Header (metadata) release May 3, 2023 - Map release May 3, 2023 - Update Jul 24, 2024 - Current status Jul 24, 2024 Processing site : PDBe / Status : Released
Show all Show less
 Yorodumi
Yorodumi Open data
Open data Basic information
Basic information
 Map data
Map data Sample
Sample Keywords
Keywords Function and homology information
Function and homology information Homo sapiens (human) /
Homo sapiens (human) / 
 unidentified adenovirus
unidentified adenovirus Authors
Authors Germany, European Union, 2 items
Germany, European Union, 2 items  Citation
Citation Journal: Mol Cell / Year: 2023
Journal: Mol Cell / Year: 2023
 Structure visualization
Structure visualization Downloads & links
Downloads & links emd_16331.map.gz
emd_16331.map.gz EMDB map data format
EMDB map data format emd-16331-v30.xml
emd-16331-v30.xml emd-16331.xml
emd-16331.xml EMDB header
EMDB header emd_16331_fsc.xml
emd_16331_fsc.xml FSC data file
FSC data file emd_16331.png
emd_16331.png emd_16331_msk_1.map
emd_16331_msk_1.map Mask map
Mask map emd-16331.cif.gz
emd-16331.cif.gz emd_16331_half_map_1.map.gz
emd_16331_half_map_1.map.gz emd_16331_half_map_2.map.gz
emd_16331_half_map_2.map.gz http://ftp.pdbj.org/pub/emdb/structures/EMD-16331
http://ftp.pdbj.org/pub/emdb/structures/EMD-16331 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-16331
ftp://ftp.pdbj.org/pub/emdb/structures/EMD-16331


 F&H Search
F&H Search Links
Links EMDB (EBI/PDBe) /
EMDB (EBI/PDBe) /  EMDataResource
EMDataResource Map
Map Download / File: emd_16331.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES)
Download / File: emd_16331.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) emd_16331_msk_1.map
emd_16331_msk_1.map Sample components
Sample components Processing
Processing Sample preparation
Sample preparation Electron microscopy
Electron microscopy FIELD EMISSION GUN
FIELD EMISSION GUN
 Image processing
Image processing
 Movie
Movie Controller
Controller














































 X (Sec.)
X (Sec.) Y (Row.)
Y (Row.) Z (Col.)
Z (Col.)












































 Trichoplusia ni (cabbage looper)
Trichoplusia ni (cabbage looper)