[English] 日本語
 Yorodumi
Yorodumi- EMDB-1184: Interactions of the release factor RF1 with the ribosome as revea... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-1184 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Interactions of the release factor RF1 with the ribosome as revealed by cryo-EM. | |||||||||
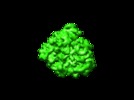
 Map data Map data | This is an em map of a 70s E.coli ribosome | |||||||||
 Sample Sample |
| |||||||||
| Function / homology |  Function and homology information Function and homology information | |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 12.8 Å | |||||||||
 Authors Authors | Rawat U / Gao H / Zavialov A / Gursky R / Ehrenberg M / Frank J | |||||||||
 Citation Citation |  Journal: J Mol Biol / Year: 2006 Journal: J Mol Biol / Year: 2006Title: Interactions of the release factor RF1 with the ribosome as revealed by cryo-EM. Authors: Urmila Rawat / Haixiao Gao / Andrey Zavialov / Richard Gursky / Måns Ehrenberg / Joachim Frank /  Abstract: In eubacteria, termination of translation is signaled by any one of the stop codons UAA, UAG, and UGA moving into the ribosomal A site. Two release factors, RF1 and RF2, recognize and bind to the ...In eubacteria, termination of translation is signaled by any one of the stop codons UAA, UAG, and UGA moving into the ribosomal A site. Two release factors, RF1 and RF2, recognize and bind to the stop codons with different affinities and trigger the hydrolysis of the ester bond that links the polypeptide with the P-site tRNA. Cryo-electron microscopy (cryo-EM) results obtained in this study show that ribosome-bound RF1 is in an open conformation, unlike the closed conformation observed in the crystal structure of the free factor, allowing its simultaneous access to both the decoding center and the peptidyl-transferase center. These results are similar to those obtained for RF2, but there is an important difference in how the factors bind to protein L11, which forms part of the GTPase-associated center of the large ribosomal subunit. The difference in the binding position, C-terminal domain for RF2 versus N-terminal domain for RF1, explains a body of L11 mutation studies that revealed differential effects on the activity of the two factors. Very recent data obtained with small-angle X-ray scattering now reveal that the solution structure of RF1 is open, as here seen on the ribosome by cryo-EM, and not closed, as seen in the crystal. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_1184.map.gz emd_1184.map.gz | 7.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-1184-v30.xml emd-1184-v30.xml emd-1184.xml emd-1184.xml | 11.6 KB 11.6 KB | Display Display |  EMDB header EMDB header |
| Images |  1184.gif 1184.gif | 11.8 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-1184 http://ftp.pdbj.org/pub/emdb/structures/EMD-1184 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-1184 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-1184 | HTTPS FTP |
-Validation report
| Summary document |  emd_1184_validation.pdf.gz emd_1184_validation.pdf.gz | 315.9 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_1184_full_validation.pdf.gz emd_1184_full_validation.pdf.gz | 315.4 KB | Display | |
| Data in XML |  emd_1184_validation.xml.gz emd_1184_validation.xml.gz | 5.7 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-1184 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-1184 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-1184 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-1184 | HTTPS FTP |
-Related structure data
| Related structure data |  2fvoMC  3dg4M  1185C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_1184.map.gz / Format: CCP4 / Size: 8.2 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_1184.map.gz / Format: CCP4 / Size: 8.2 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | This is an em map of a 70s E.coli ribosome | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2.82 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Release complex bound with RF1-WT
| Entire | Name: Release complex bound with RF1-WT |
|---|---|
| Components |
|
-Supramolecule #1000: Release complex bound with RF1-WT
| Supramolecule | Name: Release complex bound with RF1-WT / type: sample / ID: 1000 / Number unique components: 6 |
|---|
-Supramolecule #1: 30S
| Supramolecule | Name: 30S / type: complex / ID: 1 / Recombinant expression: No / Ribosome-details: ribosome-prokaryote: SSU 30S |
|---|---|
| Source (natural) | Organism:  |
-Supramolecule #2: 50S
| Supramolecule | Name: 50S / type: complex / ID: 2 / Recombinant expression: No / Ribosome-details: ribosome-prokaryote: LSU 50S |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: P-site tRNA
| Macromolecule | Name: P-site tRNA / type: rna / ID: 1 / Classification: OTHER / Structure: OTHER / Synthetic?: No |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #2: E-site tRNA
| Macromolecule | Name: E-site tRNA / type: rna / ID: 2 / Classification: OTHER / Structure: OTHER / Synthetic?: No |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #3: mRNA
| Macromolecule | Name: mRNA / type: rna / ID: 3 / Classification: OTHER / Structure: DOUBLE HELIX / Synthetic?: Yes |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
-Macromolecule #4: RF1
| Macromolecule | Name: RF1 / type: protein_or_peptide / ID: 4 / Recombinant expression: No |
|---|---|
| Source (natural) | Organism:  |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 90 % / Chamber temperature: 93 K / Instrument: HOMEMADE PLUNGER Details: Vitrification instrument: two sided blotting plunger Method: Blot for 2 seconds before plunging |
|---|
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI F20 |
|---|---|
| Temperature | Average: 93 K |
| Alignment procedure | Legacy - Electron beam tilt params: 0 |
| Date | Dec 1, 2002 |
| Image recording | Category: FILM / Film or detector model: KODAK SO-163 FILM / Digitization - Scanner: ZEISS SCAI / Digitization - Sampling interval: 14 µm / Number real images: 56 / Average electron dose: 20 e/Å2 / Od range: 1.2 / Bits/pixel: 12 |
| Tilt angle min | 0 |
| Tilt angle max | 0 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated magnification: 49696 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2 mm / Nominal defocus max: 4.0 µm / Nominal defocus min: 2.0 µm / Nominal magnification: 50000 |
| Sample stage | Specimen holder: Cryo stage / Specimen holder model: GATAN LIQUID NITROGEN |
| Experimental equipment |  Model: Tecnai F20 / Image courtesy: FEI Company |
- Image processing
Image processing
| CTF correction | Details: defocus groups |
|---|---|
| Final reconstruction | Applied symmetry - Point group: C1 (asymmetric) / Algorithm: OTHER / Resolution.type: BY AUTHOR / Resolution: 12.8 Å / Resolution method: FSC 0.5 CUT-OFF / Software - Name: SPIDER, package / Number images used: 24622 |
| Final angle assignment | Details: SPIDER: theta 15 degrees, phi 15 degrees |
-Atomic model buiding 1
| Initial model | PDB ID: Chain - Chain ID: A |
|---|---|
| Software | Name: O |
| Details | Protocol: Rigid Body. manual fitting in O |
| Refinement | Space: REAL / Protocol: RIGID BODY FIT / Target criteria: correlation coefficient |
| Output model |  PDB-2fvo:  PDB-3dg4: |
 Movie
Movie Controller
Controller













 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)






















