[English] 日本語
 Yorodumi
Yorodumi- EMDB-1302: RF3 induces ribosomal conformational changes responsible for diss... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-1302 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | RF3 induces ribosomal conformational changes responsible for dissociation of class I release factors. | |||||||||
 Map data Map data | This is the map of a ribosome. | |||||||||
 Sample Sample |
| |||||||||
| Function / homology |  Function and homology information Function and homology informationregulation of translational termination / translation release factor activity, codon nonspecific / translation release factor activity, codon specific / guanosine tetraphosphate binding / translational termination / maintenance of translational fidelity / GDP binding / GTPase activity / GTP binding / cytoplasm / cytosol Similarity search - Function | |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / negative staining / Resolution: 15.5 Å | |||||||||
 Authors Authors | Gao H / Zhou Z / Rawat U / Huang C / Bouakaz L / Wang C / Liu Y / Zavialov A / Gursky R / Sanyal S ...Gao H / Zhou Z / Rawat U / Huang C / Bouakaz L / Wang C / Liu Y / Zavialov A / Gursky R / Sanyal S / Ehrenberg M / Frank J / Song H | |||||||||
 Citation Citation |  Journal: Cell / Year: 2007 Journal: Cell / Year: 2007Title: RF3 induces ribosomal conformational changes responsible for dissociation of class I release factors. Authors: Haixiao Gao / Zhihong Zhou / Urmila Rawat / Chenhui Huang / Lamine Bouakaz / Chernhoe Wang / Zhihong Cheng / Yuying Liu / Andrey Zavialov / Richard Gursky / Suparna Sanyal / Måns Ehrenberg ...Authors: Haixiao Gao / Zhihong Zhou / Urmila Rawat / Chenhui Huang / Lamine Bouakaz / Chernhoe Wang / Zhihong Cheng / Yuying Liu / Andrey Zavialov / Richard Gursky / Suparna Sanyal / Måns Ehrenberg / Joachim Frank / Haiwei Song /  Abstract: During translation termination, class II release factor RF3 binds to the ribosome to promote rapid dissociation of a class I release factor (RF) in a GTP-dependent manner. We present the crystal ...During translation termination, class II release factor RF3 binds to the ribosome to promote rapid dissociation of a class I release factor (RF) in a GTP-dependent manner. We present the crystal structure of E. coli RF3*GDP, which has a three-domain architecture strikingly similar to the structure of EF-Tu*GTP. Biochemical data on RF3 mutants show that a surface region involving domains II and III is important for distinct steps in the action cycle of RF3. Furthermore, we present a cryo-electron microscopy (cryo-EM) structure of the posttermination ribosome bound with RF3 in the GTP form. Our data show that RF3*GTP binding induces large conformational changes in the ribosome, which break the interactions of the class I RF with both the decoding center and the GTPase-associated center of the ribosome, apparently leading to the release of the class I RF. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_1302.map.gz emd_1302.map.gz | 7.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-1302-v30.xml emd-1302-v30.xml emd-1302.xml emd-1302.xml | 11.9 KB 11.9 KB | Display Display |  EMDB header EMDB header |
| Images |  1302.gif 1302.gif | 48.2 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-1302 http://ftp.pdbj.org/pub/emdb/structures/EMD-1302 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-1302 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-1302 | HTTPS FTP |
-Related structure data
| Related structure data |  2o0fMC  3dg5M  2h5eC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_1302.map.gz / Format: CCP4 / Size: 8.2 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_1302.map.gz / Format: CCP4 / Size: 8.2 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | This is the map of a ribosome. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2.82 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
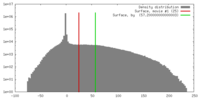
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : E.coli 70s ribosomal release complex bound with RF3
| Entire | Name: E.coli 70s ribosomal release complex bound with RF3 |
|---|---|
| Components |
|
-Supramolecule #1000: E.coli 70s ribosomal release complex bound with RF3
| Supramolecule | Name: E.coli 70s ribosomal release complex bound with RF3 / type: sample / ID: 1000 / Number unique components: 3 |
|---|
-Supramolecule #1: 70s
| Supramolecule | Name: 70s / type: complex / ID: 1 / Details: E.coli / Ribosome-details: ribosome-prokaryote: ALL |
|---|
-Macromolecule #1: RF3
| Macromolecule | Name: RF3 / type: protein_or_peptide / ID: 1 / Details: E.coli / Number of copies: 1 / Oligomeric state: Monomer / Recombinant expression: No |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #2: tRNA
| Macromolecule | Name: tRNA / type: rna / ID: 2 / Details: Hybrid P/E / Classification: OTHER / Structure: OTHER / Synthetic?: No |
|---|---|
| Source (natural) | Organism:  |
-Experimental details
-Structure determination
| Method | negative staining, cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 / Details: Polymix |
|---|---|
| Staining | Type: NEGATIVE / Details: Cryo |
| Grid | Details: Quanti-foil grids coated with a thin carbon layer |
| Vitrification | Cryogen name: ETHANE / Instrument: OTHER Details: Vitrification instrument: Vitrobot. Rapid-freezing in liquid ethane |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI F20 |
|---|---|
| Temperature | Average: 93 K |
| Alignment procedure | Legacy - Astigmatism: objective lens astigmatism was corrected at 100,000 times magnification Legacy - Electron beam tilt params: 0 |
| Date | Jan 30, 2002 |
| Image recording | Category: FILM / Film or detector model: KODAK SO-163 FILM / Digitization - Scanner: ZEISS SCAI / Digitization - Sampling interval: 14 µm / Average electron dose: 20 e/Å2 |
| Tilt angle min | 0 |
| Tilt angle max | 0 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated magnification: 49696 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.0 mm / Nominal defocus max: 4.0 µm / Nominal defocus min: 2.0 µm / Nominal magnification: 50000 |
| Sample stage | Specimen holder: cryo transfer / Specimen holder model: GATAN LIQUID NITROGEN |
| Experimental equipment |  Model: Tecnai F20 / Image courtesy: FEI Company |
- Image processing
Image processing
| CTF correction | Details: CTF correctionn of 3D map |
|---|---|
| Final reconstruction | Applied symmetry - Point group: C1 (asymmetric) / Algorithm: OTHER / Resolution.type: BY AUTHOR / Resolution: 15.5 Å / Resolution method: FSC 0.5 CUT-OFF / Software - Name: SPIDER package / Number images used: 45000 |
-Atomic model buiding 1
| Initial model | PDB ID:  2avy Chain - Chain ID: A |
|---|---|
| Software | Name: RSRef, TNT |
| Details | Protocol: Rigid Body. Real-space refinement using RSRef. |
| Refinement | Space: REAL / Protocol: RIGID BODY FIT / Target criteria: cross correlation coefficient |
| Output model |  PDB-2o0f:  PDB-3dg5: |
-Atomic model buiding 2
| Initial model | PDB ID:  2aw4 Chain - Chain ID: A |
|---|---|
| Software | Name: RSRef, TNT |
| Details | Protocol: Rigid Body. Real-space refinement using RSRef. |
| Refinement | Space: REAL / Protocol: RIGID BODY FIT / Target criteria: cross correlation coefficient |
| Output model |  PDB-2o0f:  PDB-3dg5: |
 Movie
Movie Controller
Controller


















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)