+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-11516 | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | LH2 complex from Marichromatium purpuratum | ||||||||||||
 Map data Map data | A cryo-EM map of the light harvesting complex 2 (LH2) from a photosynthetic bacterium Marichromatium purpuratum. | ||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | photosynthesis / purple bacteria / light harvesting complex / Marichromatium purpuratum / okenone. | ||||||||||||
| Function / homology |  Function and homology information Function and homology informationlight-harvesting complex / plasma membrane light-harvesting complex / bacteriochlorophyll binding / photosynthesis, light reaction / metal ion binding / plasma membrane Similarity search - Function | ||||||||||||
| Biological species |  Marichromatium purpuratum (bacteria) / Marichromatium purpuratum (bacteria) /  Marichromatium purpuratum 984 (bacteria) Marichromatium purpuratum 984 (bacteria) | ||||||||||||
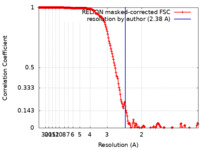
| Method | single particle reconstruction / cryo EM / Resolution: 2.38 Å | ||||||||||||
 Authors Authors | Gardiner AT / Naydenova K | ||||||||||||
| Funding support |  United States, United States,  United Kingdom, 3 items United Kingdom, 3 items
| ||||||||||||
 Citation Citation |  Journal: Sci Adv / Year: 2021 Journal: Sci Adv / Year: 2021Title: The 2.4 Å cryo-EM structure of a heptameric light-harvesting 2 complex reveals two carotenoid energy transfer pathways. Authors: Alastair T Gardiner / Katerina Naydenova / Pablo Castro-Hartmann / Tu C Nguyen-Phan / Christopher J Russo / Kasim Sader / C Neil Hunter / Richard J Cogdell / Pu Qian /   Abstract: We report the 2.4 Ångström resolution structure of the light-harvesting 2 (LH2) complex from () determined by cryogenic electron microscopy. The structure contains a heptameric ring that is ...We report the 2.4 Ångström resolution structure of the light-harvesting 2 (LH2) complex from () determined by cryogenic electron microscopy. The structure contains a heptameric ring that is unique among all known LH2 structures, explaining the unusual spectroscopic properties of this bacterial antenna complex. We identify two sets of distinct carotenoids in the structure and describe a network of energy transfer pathways from the carotenoids to bacteriochlorophyll molecules. The geometry imposed by the heptameric ring controls the resonant coupling of the long-wavelength energy absorption band. Together, these details reveal key aspects of the assembly and oligomeric form of purple bacterial LH2 complexes that were previously inaccessible by any technique. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_11516.map.gz emd_11516.map.gz | 32 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-11516-v30.xml emd-11516-v30.xml emd-11516.xml emd-11516.xml | 17.3 KB 17.3 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_11516_fsc.xml emd_11516_fsc.xml | 18.1 KB | Display |  FSC data file FSC data file |
| Images |  emd_11516.png emd_11516.png | 189.7 KB | ||
| Filedesc metadata |  emd-11516.cif.gz emd-11516.cif.gz | 6.1 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-11516 http://ftp.pdbj.org/pub/emdb/structures/EMD-11516 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-11516 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-11516 | HTTPS FTP |
-Related structure data
| Related structure data |  6zxaMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | |
| EM raw data |  EMPIAR-10606 (Title: Movies of a heptameric light-harvesting 2 complex from the marine purple bacterium Mch. purpuratum EMPIAR-10606 (Title: Movies of a heptameric light-harvesting 2 complex from the marine purple bacterium Mch. purpuratumData size: 12.2 TB Data #1: Unaligned multi-frame micrographs of purple bacterial LH2 on HexAuFoil supports [micrographs - multiframe]) |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_11516.map.gz / Format: CCP4 / Size: 512 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_11516.map.gz / Format: CCP4 / Size: 512 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | A cryo-EM map of the light harvesting complex 2 (LH2) from a photosynthetic bacterium Marichromatium purpuratum. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.646 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Light harvesting complex 2 (LH2)
| Entire | Name: Light harvesting complex 2 (LH2) |
|---|---|
| Components |
|
-Supramolecule #1: Light harvesting complex 2 (LH2)
| Supramolecule | Name: Light harvesting complex 2 (LH2) / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 Details: The LH2 complex from Mar. purpuratum forms a circular heptameter. Each subunit consists of an alpha/beta pair, in which all pigment molecules, three Bchl a and two carotenoids are bound non-covalently. |
|---|---|
| Source (natural) | Organism:  Marichromatium purpuratum (bacteria) / Strain: BN5500 Marichromatium purpuratum (bacteria) / Strain: BN5500 |
| Molecular weight | Theoretical: 110 KDa |
-Macromolecule #1: LHC domain-containing protein
| Macromolecule | Name: LHC domain-containing protein / type: protein_or_peptide / ID: 1 / Number of copies: 7 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Marichromatium purpuratum 984 (bacteria) Marichromatium purpuratum 984 (bacteria) |
| Molecular weight | Theoretical: 8.015369 KDa |
| Sequence | String: MKVPVMMADE SIATINHPED DWKIWTVINP ATWMVPFFGI LFVQMWLIHS YALSLPGYGF KDSVRVAQPA UniProtKB: Uncharacterized protein |
-Macromolecule #2: Light-harvesting protein B:800-850 subunit beta
| Macromolecule | Name: Light-harvesting protein B:800-850 subunit beta / type: protein_or_peptide / ID: 2 / Number of copies: 7 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Marichromatium purpuratum 984 (bacteria) Marichromatium purpuratum 984 (bacteria) |
| Molecular weight | Theoretical: 5.554275 KDa |
| Sequence | String: ADPKAANLSG LTDAQAKEFH EHWKHGVWSW VMIASAVHVV TWIYQPWF UniProtKB: Light-harvesting protein B:800-850 subunit beta |
-Macromolecule #3: BACTERIOCHLOROPHYLL A
| Macromolecule | Name: BACTERIOCHLOROPHYLL A / type: ligand / ID: 3 / Number of copies: 21 / Formula: BCL |
|---|---|
| Molecular weight | Theoretical: 911.504 Da |
| Chemical component information |  ChemComp-BCL: |
-Macromolecule #4: 9-cis-okenone
| Macromolecule | Name: 9-cis-okenone / type: ligand / ID: 4 / Number of copies: 7 / Formula: QS2 |
|---|---|
| Molecular weight | Theoretical: 578.866 Da |
| Chemical component information | 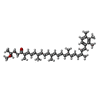 ChemComp-QS2: |
-Macromolecule #5: all-trans okenone
| Macromolecule | Name: all-trans okenone / type: ligand / ID: 5 / Number of copies: 7 / Formula: QSE |
|---|---|
| Molecular weight | Theoretical: 578.866 Da |
| Chemical component information | 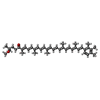 ChemComp-QSE: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 5.5 mg/mL |
|---|---|
| Buffer | pH: 8 / Component - Concentration: 20.0 mM/L / Component - Name: Tris.HCl Details: The final buffer contains 0.02 % n-dodecyl-beta-D-maltopyranoside (DDM) |
| Grid | Model: Homemade / Material: GOLD / Mesh: 400 / Pretreatment - Type: PLASMA CLEANING / Pretreatment - Time: 60 sec. / Pretreatment - Atmosphere: OTHER / Pretreatment - Pressure: 0.0067 kPa / Details: Fischione 1070 |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: HOMEMADE PLUNGER / Details: Blowing time, 10.0 sec.. |
| Details | The sample was mono disperse with detergent beta-DDM. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Temperature | Min: 80.0 K |
| Image recording | Film or detector model: FEI FALCON IV (4k x 4k) / Number grids imaged: 1 / Number real images: 9543 / Average exposure time: 0.29 sec. / Average electron dose: 43.7 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Calibrated defocus max: 3.0 µm / Calibrated defocus min: 0.4 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.4 µm / Nominal defocus min: 0.8 µm / Nominal magnification: 120000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model |
| ||||||
|---|---|---|---|---|---|---|---|
| Refinement | Space: REAL / Protocol: FLEXIBLE FIT / Overall B value: 22.55 | ||||||
| Output model |  PDB-6zxa: |
 Movie
Movie Controller
Controller












 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)