[English] 日本語
 Yorodumi
Yorodumi- EMDB-10911: Cryo-EM structure of T7 bacteriophage DNA translocation gp15 core... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-10911 | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of T7 bacteriophage DNA translocation gp15 core protein intermediate assembly | ||||||||||||
 Map data Map data | T7 DNA ejection complex | ||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | VIRAL PROTEIN / DNA TRANSLOCATION / VIRAL INFECTION / PERIPLASMIC SPACE COMPLEX. | ||||||||||||
| Function / homology | Internal virion protein Gp15 / host cell periplasmic space / symbiont genome ejection through host cell envelope, short tail mechanism / virion component / Internal virion protein gp15 Function and homology information Function and homology information | ||||||||||||
| Biological species |   Escherichia phage T7 (virus) Escherichia phage T7 (virus) | ||||||||||||
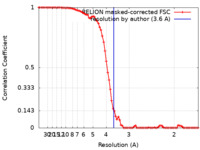
| Method | single particle reconstruction / cryo EM / Resolution: 3.6 Å | ||||||||||||
 Authors Authors | Perez-Ruiz M / Pulido-Cid M | ||||||||||||
| Funding support |  Spain, 3 items Spain, 3 items
| ||||||||||||
 Citation Citation |  Journal: Proc Natl Acad Sci U S A / Year: 2021 Journal: Proc Natl Acad Sci U S A / Year: 2021Title: Assisted assembly of bacteriophage T7 core components for genome translocation across the bacterial envelope. Authors: Mar Pérez-Ruiz / Mar Pulido-Cid / Juan Román Luque-Ortega / José María Valpuesta / Ana Cuervo / José L Carrascosa /  Abstract: In most bacteriophages, genome transport across bacterial envelopes is carried out by the tail machinery. In viruses of the family, in which the tail is not long enough to traverse the bacterial ...In most bacteriophages, genome transport across bacterial envelopes is carried out by the tail machinery. In viruses of the family, in which the tail is not long enough to traverse the bacterial wall, it has been postulated that viral core proteins assembled inside the viral head are translocated and reassembled into a tube within the periplasm that extends the tail channel. Bacteriophage T7 infects , and despite extensive studies, the precise mechanism by which its genome is translocated remains unknown. Using cryo-electron microscopy, we have resolved the structure of two different assemblies of the T7 DNA translocation complex composed of the core proteins gp15 and gp16. Gp15 alone forms a partially folded hexamer, which is further assembled upon interaction with gp16 into a tubular structure, forming a channel that could allow DNA passage. The structure of the gp15-gp16 complex also shows the location within gp16 of a canonical transglycosylase motif involved in the degradation of the bacterial peptidoglycan layer. This complex docks well in the tail extension structure found in the periplasm of T7-infected bacteria and matches the sixfold symmetry of the phage tail. In such cases, gp15 and gp16 that are initially present in the T7 capsid eightfold-symmetric core would change their oligomeric state upon reassembly in the periplasm. Altogether, these results allow us to propose a model for the assembly of the core translocation complex in the periplasm, which furthers understanding of the molecular mechanism involved in the release of T7 viral DNA into the bacterial cytoplasm. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_10911.map.gz emd_10911.map.gz | 38.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-10911-v30.xml emd-10911-v30.xml emd-10911.xml emd-10911.xml | 13.5 KB 13.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_10911_fsc.xml emd_10911_fsc.xml | 8.2 KB | Display |  FSC data file FSC data file |
| Images |  emd_10911.png emd_10911.png | 71.8 KB | ||
| Filedesc metadata |  emd-10911.cif.gz emd-10911.cif.gz | 6 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-10911 http://ftp.pdbj.org/pub/emdb/structures/EMD-10911 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-10911 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-10911 | HTTPS FTP |
-Validation report
| Summary document |  emd_10911_validation.pdf.gz emd_10911_validation.pdf.gz | 475.3 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_10911_full_validation.pdf.gz emd_10911_full_validation.pdf.gz | 474.8 KB | Display | |
| Data in XML |  emd_10911_validation.xml.gz emd_10911_validation.xml.gz | 10.2 KB | Display | |
| Data in CIF |  emd_10911_validation.cif.gz emd_10911_validation.cif.gz | 13.4 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-10911 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-10911 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-10911 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-10911 | HTTPS FTP |
-Related structure data
| Related structure data |  6yszMC  6yt5C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_10911.map.gz / Format: CCP4 / Size: 52.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_10911.map.gz / Format: CCP4 / Size: 52.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | T7 DNA ejection complex | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.85 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : gp15 tubular core protein
| Entire | Name: gp15 tubular core protein |
|---|---|
| Components |
|
-Supramolecule #1: gp15 tubular core protein
| Supramolecule | Name: gp15 tubular core protein / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:   Escherichia phage T7 (virus) Escherichia phage T7 (virus) |
| Molecular weight | Theoretical: 543 KDa |
-Macromolecule #1: Internal virion protein gp15
| Macromolecule | Name: Internal virion protein gp15 / type: protein_or_peptide / ID: 1 / Number of copies: 6 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Escherichia phage T7 (virus) Escherichia phage T7 (virus) |
| Molecular weight | Theoretical: 88.377219 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MRGSHHHHHH GMASMTGGQQ MGRDLYDDDD KDPSSMSKIE SALQAAQPGL SRLRGGAGGM GYRAATTQAE QPRSSLLDTI GRFAKAGAD MYTAKEQRAR DLADERSNEI IRKLTPEQRR EALNNGTLLY QDDPYAMEAL RVKTGRNAAY LVDDDVMQKI K EGVFRTRE ...String: MRGSHHHHHH GMASMTGGQQ MGRDLYDDDD KDPSSMSKIE SALQAAQPGL SRLRGGAGGM GYRAATTQAE QPRSSLLDTI GRFAKAGAD MYTAKEQRAR DLADERSNEI IRKLTPEQRR EALNNGTLLY QDDPYAMEAL RVKTGRNAAY LVDDDVMQKI K EGVFRTRE EMEEYRHSRL QEGAKVYAEQ FGIDPEDVDY QRGFNGDITE RNISLYGAHD NFLSQQAQKG AIMNSRVELN GV LQDPDML RRPDSADFFE KYIDNGLVTG AIPSDAQATQ LISQAFSDAS SRAGGADFLM RVGDKKVTLN GATTTYRELI GEE QWNALM VTAQRSQFET DAKLNEQYRL KINSALNQED PRTAWEMLQG IKAELDKVQP DEQMTPQREW LISAQEQVQN QMNA WTKAQ AKALDDSMKS MNKLDVIDKQ FQKRINGEWV STDFKDMPVN ENTGEFKHSD MVNYANKKLA EIDSMDIPDG AKDAM KLKY LQADSKDGAF RTAIGTMVTD AGQEWSAAVI NGKLPERTPA MDALRRIRNA DPQLIAALYP DQAELFLTMD MMDKQG IDP QVILDADRLT VKRSKEQRFE DDKAFESALN ASKAPEIARM PASLRESARK IYDSVKYRSG NESMAMEQMT KFLKEST YT FTGDDVDGDT VGVIPKNMMQ VNSDPKSWEQ GRDILEEARK GIIASNPWIT NKQLTMYSQG DSIYLMDTTG QVRVRYDK E LLSKVWSENQ KKLEEKAREK ALADVNKRAP IVAATKAREA AAKRVREKRK QTPKFIYGRK E UniProtKB: Internal virion protein gp15 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1 mg/mL | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 8 Component:
| ||||||||
| Grid | Model: Quantifoil R2/2 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 60 sec. / Pretreatment - Atmosphere: AIR | ||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 95 % / Chamber temperature: 293.15 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TALOS ARCTICA |
|---|---|
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Detector mode: COUNTING / Number grids imaged: 1 / Number real images: 2470 / Average exposure time: 35.0 sec. / Average electron dose: 30.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated magnification: 120000 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: AB INITIO MODEL |
|---|---|
| Output model |  PDB-6ysz: |
 Movie
Movie Controller
Controller












 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)