+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-10414 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | SAGA Core module | |||||||||
 Map data Map data | local resolution filtered map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Coactivator / Transcription / Histone acetyltransferase / Histone deubiquitinase / GENE REGULATION | |||||||||
| Function / homology |  Function and homology information Function and homology informationconjugation with cellular fusion / DUBm complex / RITS complex assembly / pseudohyphal growth / positive regulation of DNA-templated transcription initiation / invasive growth in response to glucose limitation / regulation of nucleocytoplasmic transport / SLIK (SAGA-like) complex / regulation of protein localization to chromatin / SAGA complex ...conjugation with cellular fusion / DUBm complex / RITS complex assembly / pseudohyphal growth / positive regulation of DNA-templated transcription initiation / invasive growth in response to glucose limitation / regulation of nucleocytoplasmic transport / SLIK (SAGA-like) complex / regulation of protein localization to chromatin / SAGA complex / RNA polymerase II transcribes snRNA genes / RNA Polymerase II Promoter Escape / RNA Polymerase II Transcription Pre-Initiation And Promoter Opening / RNA Polymerase II Transcription Initiation / RNA Polymerase II Transcription Initiation And Promoter Clearance / IRE1-mediated unfolded protein response / RNA polymerase II general transcription initiation factor activity / RNA Polymerase II Pre-transcription Events / transcription factor TFIID complex / positive regulation of RNA polymerase II transcription preinitiation complex assembly / Ub-specific processing proteases / mRNA export from nucleus / RNA polymerase II preinitiation complex assembly / TBP-class protein binding / ubiquitin binding / transcription coregulator activity / transcription initiation at RNA polymerase II promoter / promoter-specific chromatin binding / enzyme activator activity / peroxisome / chromatin organization / protein-containing complex assembly / molecular adaptor activity / transcription by RNA polymerase II / RNA polymerase II-specific DNA-binding transcription factor binding / transcription coactivator activity / chromatin remodeling / protein heterodimerization activity / chromatin binding / regulation of transcription by RNA polymerase II / structural molecule activity / positive regulation of transcription by RNA polymerase II / mitochondrion / DNA binding / identical protein binding / nucleus / cytosol Similarity search - Function | |||||||||
| Biological species |   | |||||||||
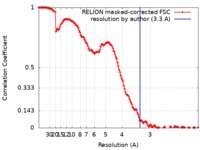
| Method | single particle reconstruction / cryo EM / Resolution: 3.3 Å | |||||||||
 Authors Authors | Wang H / Cheung A | |||||||||
| Funding support |  Germany, 2 items Germany, 2 items
| |||||||||
 Citation Citation |  Journal: Nature / Year: 2020 Journal: Nature / Year: 2020Title: Structure of the transcription coactivator SAGA. Authors: Haibo Wang / Christian Dienemann / Alexandra Stützer / Henning Urlaub / Alan C M Cheung / Patrick Cramer /   Abstract: Gene transcription by RNA polymerase II is regulated by activator proteins that recruit the coactivator complexes SAGA (Spt-Ada-Gcn5-acetyltransferase) and transcription factor IID (TFIID). SAGA is ...Gene transcription by RNA polymerase II is regulated by activator proteins that recruit the coactivator complexes SAGA (Spt-Ada-Gcn5-acetyltransferase) and transcription factor IID (TFIID). SAGA is required for all regulated transcription and is conserved among eukaryotes. SAGA contains four modules: the activator-binding Tra1 module, the core module, the histone acetyltransferase (HAT) module and the histone deubiquitination (DUB) module. Previous studies provided partial structures, but the structure of the central core module is unknown. Here we present the cryo-electron microscopy structure of SAGA from the yeast Saccharomyces cerevisiae and resolve the core module at 3.3 Å resolution. The core module consists of subunits Taf5, Sgf73 and Spt20, and a histone octamer-like fold. The octamer-like fold comprises the heterodimers Taf6-Taf9, Taf10-Spt7 and Taf12-Ada1, and two histone-fold domains in Spt3. Spt3 and the adjacent subunit Spt8 interact with the TATA box-binding protein (TBP). The octamer-like fold and its TBP-interacting region are similar in TFIID, whereas Taf5 and the Taf6 HEAT domain adopt distinct conformations. Taf12 and Spt20 form flexible connections to the Tra1 module, whereas Sgf73 tethers the DUB module. Binding of a nucleosome to SAGA displaces the HAT and DUB modules from the core-module surface, allowing the DUB module to bind one face of an ubiquitinated nucleosome. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_10414.map.gz emd_10414.map.gz | 142.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-10414-v30.xml emd-10414-v30.xml emd-10414.xml emd-10414.xml | 30.8 KB 30.8 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_10414_fsc.xml emd_10414_fsc.xml | 14.2 KB | Display |  FSC data file FSC data file |
| Images |  emd_10414.png emd_10414.png | 154.3 KB | ||
| Filedesc metadata |  emd-10414.cif.gz emd-10414.cif.gz | 9.4 KB | ||
| Others |  emd_10414_half_map_1.map.gz emd_10414_half_map_1.map.gz emd_10414_half_map_2.map.gz emd_10414_half_map_2.map.gz | 194.7 MB 194.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-10414 http://ftp.pdbj.org/pub/emdb/structures/EMD-10414 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-10414 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-10414 | HTTPS FTP |
-Related structure data
| Related structure data |  6t9kMC  6t9iC  6t9jC  6t9lC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_10414.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_10414.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | local resolution filtered map | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.05 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
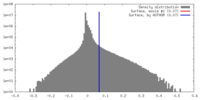
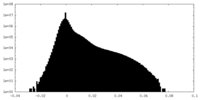
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Half map: #1
| File | emd_10414_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
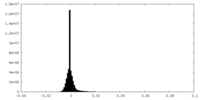
| Density Histograms |
-Half map: #2
| File | emd_10414_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
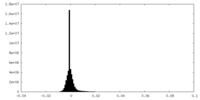
| Density Histograms |
- Sample components
Sample components
+Entire : SAGA Core module
+Supramolecule #1: SAGA Core module
+Macromolecule #1: Transcription factor SPT20
+Macromolecule #2: Protein SPT3
+Macromolecule #3: Transcription initiation factor TFIID subunit 5
+Macromolecule #4: Transcription initiation factor TFIID subunit 6
+Macromolecule #5: Transcription initiation factor TFIID subunit 9
+Macromolecule #6: Transcription initiation factor TFIID subunit 10
+Macromolecule #7: Transcriptional coactivator HFI1/ADA1
+Macromolecule #8: Transcription initiation factor TFIID subunit 12
+Macromolecule #9: Transcriptional activator SPT7
+Macromolecule #10: SAGA-associated factor 73
+Macromolecule #11: unassigned sequence
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.2 mg/mL | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
Details: Solution were made from stock solution | ||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV / Details: blot for 4 seconds before plunging. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Average exposure time: 9.0 sec. / Average electron dose: 42.45 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 70.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: AB INITIO MODEL / Overall B value: 107.1 / Target criteria: Correlation coefficient |
|---|---|
| Output model |  PDB-6t9k: |
 Movie
Movie Controller
Controller

























 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)