+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-10121 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of LptB2FGC in complex with AMP-PNP | |||||||||
 Map data Map data | None | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | lipopolysaccharide transporter / LPS / LptB2FGC / LptB / LptBFG / outer membrane / Gram-negative bacteria / ABC transporter / Inner membrane protein complex / TRANSPORT PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationTranslocases; Catalysing the translocation of carbohydrates and their derivatives; Linked to the hydrolysis of a nucleoside triphosphate / lipopolysaccharide transport / ATP-binding cassette (ABC) transporter complex / transmembrane transport / ATP hydrolysis activity / ATP binding / cytoplasm Similarity search - Function | |||||||||
| Biological species |  Shigella flexneri (bacteria) Shigella flexneri (bacteria) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.5 Å | |||||||||
 Authors Authors | Tang XD / Chang SH | |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2019 Journal: Nat Commun / Year: 2019Title: Cryo-EM structures of lipopolysaccharide transporter LptBFGC in lipopolysaccharide or AMP-PNP-bound states reveal its transport mechanism. Authors: Xiaodi Tang / Shenghai Chang / Qinghua Luo / Zhengyu Zhang / Wen Qiao / Caihuang Xu / Changbin Zhang / Yang Niu / Wenxian Yang / Ting Wang / Zhibo Zhang / Xiaofeng Zhu / Xiawei Wei / ...Authors: Xiaodi Tang / Shenghai Chang / Qinghua Luo / Zhengyu Zhang / Wen Qiao / Caihuang Xu / Changbin Zhang / Yang Niu / Wenxian Yang / Ting Wang / Zhibo Zhang / Xiaofeng Zhu / Xiawei Wei / Changjiang Dong / Xing Zhang / Haohao Dong /   Abstract: Lipopolysaccharides (LPS) of Gram-negative bacteria are critical for the defence against cytotoxic substances and must be transported from the inner membrane (IM) to the outer membrane (OM) through ...Lipopolysaccharides (LPS) of Gram-negative bacteria are critical for the defence against cytotoxic substances and must be transported from the inner membrane (IM) to the outer membrane (OM) through a bridge formed by seven membrane proteins (LptBFGCADE). The IM component LptBFG powers the process through a yet unclarified mechanism. Here we report three high-resolution cryo-EM structures of LptBFG alone and complexed with LptC (LptBFGC), trapped in either the LPS- or AMP-PNP-bound state. The structures reveal conformational changes between these states and substrate binding with or without LptC. We identify two functional transmembrane arginine-containing loops interacting with the bound AMP-PNP and elucidate allosteric communications between the domains. AMP-PNP binding induces an inward rotation and shift of the transmembrane helices of LptFG and LptC to tighten the cavity, with the closure of two lateral gates, to eventually expel LPS into the bridge. Functional assays reveal the functionality of the LptF and LptG periplasmic domains. Our findings shed light on the LPS transport mechanism. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_10121.map.gz emd_10121.map.gz | 20.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-10121-v30.xml emd-10121-v30.xml emd-10121.xml emd-10121.xml | 16 KB 16 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_10121.png emd_10121.png | 54.3 KB | ||
| Filedesc metadata |  emd-10121.cif.gz emd-10121.cif.gz | 6.2 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-10121 http://ftp.pdbj.org/pub/emdb/structures/EMD-10121 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-10121 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-10121 | HTTPS FTP |
-Validation report
| Summary document |  emd_10121_validation.pdf.gz emd_10121_validation.pdf.gz | 514.3 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_10121_full_validation.pdf.gz emd_10121_full_validation.pdf.gz | 513.8 KB | Display | |
| Data in XML |  emd_10121_validation.xml.gz emd_10121_validation.xml.gz | 5.6 KB | Display | |
| Data in CIF |  emd_10121_validation.cif.gz emd_10121_validation.cif.gz | 6.3 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-10121 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-10121 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-10121 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-10121 | HTTPS FTP |
-Related structure data
| Related structure data |  6s8gMC  6s8hC  6s8nC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_10121.map.gz / Format: CCP4 / Size: 22.2 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_10121.map.gz / Format: CCP4 / Size: 22.2 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | None | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.014 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : LptB2FGC
| Entire | Name: LptB2FGC |
|---|---|
| Components |
|
-Supramolecule #1: LptB2FGC
| Supramolecule | Name: LptB2FGC / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#3 / Details: LPS transporter LptB2FGC |
|---|---|
| Source (natural) | Organism:  Shigella flexneri (bacteria) Shigella flexneri (bacteria) |
-Macromolecule #1: Lipopolysaccharide ABC transporter, ATP-binding protein LptB
| Macromolecule | Name: Lipopolysaccharide ABC transporter, ATP-binding protein LptB type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Shigella flexneri (bacteria) Shigella flexneri (bacteria) |
| Molecular weight | Theoretical: 26.837668 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MATLTAKNLA KAYKGRRVVE DVSLTVNSGE IVGLLGPNGA GKTTTFYMVV GIVPRDAGNI IIDDDDISLL PLHARARRGI GYLPQEASI FRRLSVYDNL MAVLQIRDDL SAEQREDRAN ELMEEFHIEH LRDSMGQSLS GGERRRVEIA RALAANPKFI L LDEPFAGV ...String: MATLTAKNLA KAYKGRRVVE DVSLTVNSGE IVGLLGPNGA GKTTTFYMVV GIVPRDAGNI IIDDDDISLL PLHARARRGI GYLPQEASI FRRLSVYDNL MAVLQIRDDL SAEQREDRAN ELMEEFHIEH LRDSMGQSLS GGERRRVEIA RALAANPKFI L LDEPFAGV DPISVIDIKR IIEHLRDSGL GVLITDHNVR ETLAVCERAY IVSQGHLIAH GTPTEILQDE HVKRVYLGED FR L UniProtKB: Lipopolysaccharide export system ATP-binding protein LptB |
-Macromolecule #2: LPS export ABC transporter permease LptF
| Macromolecule | Name: LPS export ABC transporter permease LptF / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Shigella flexneri (bacteria) Shigella flexneri (bacteria) |
| Molecular weight | Theoretical: 40.45352 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MIIIRYLVRE TLKSQLAILF ILLLIFFCQK LVRILGAAVD GDIPANLVLS LLGLGVPEMA QLILPLSLFL GLLMTLGKLY TESEITVMH ACGLSKAVLV KAAMILAVFT AIVAAVNVMW AGPWSSRHQD EVLEEAKANP GMAALAQGQF QQATNGSSVL F IESVDGSD ...String: MIIIRYLVRE TLKSQLAILF ILLLIFFCQK LVRILGAAVD GDIPANLVLS LLGLGVPEMA QLILPLSLFL GLLMTLGKLY TESEITVMH ACGLSKAVLV KAAMILAVFT AIVAAVNVMW AGPWSSRHQD EVLEEAKANP GMAALAQGQF QQATNGSSVL F IESVDGSD FKDVFLAQIR PKGNARPSVV VADSGHLTQL RDGSQVVTLN QGTRFEGTAL LRDFRITDFQ DYQAIIGHQA VA LDPNDTD QMDMRTLWNT DTDRARAELN WRITLVVTVF MMALMVVPLS VVNPRQGRVL SMLPAMLLYL LFFLIQTSLK SNG GKGKLD PTLWMWTVNL IYLALAIVLN LWDTVFVRRL RASFSRKGAV UniProtKB: Lipopolysaccharide export system permease protein LptF |
-Macromolecule #3: Inner membrane protein yjgQ
| Macromolecule | Name: Inner membrane protein yjgQ / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Shigella flexneri (bacteria) Shigella flexneri (bacteria) |
| Molecular weight | Theoretical: 39.622414 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MQPFGVLDRY IGKTIFTTIM MTLFMLVSLS GIIKFVDQLK KAGQGSYDAL GAGMYTLLSV PKDVQIFFPM AALLGALLGL GMLAQRSEL VVMQASGFTR MQVALSVMKT AIPLVLLTMA IGEWVAPQGE QMARNYRAQA MYGGSLLSTQ QGLWAKDGNN F VYIERVKG ...String: MQPFGVLDRY IGKTIFTTIM MTLFMLVSLS GIIKFVDQLK KAGQGSYDAL GAGMYTLLSV PKDVQIFFPM AALLGALLGL GMLAQRSEL VVMQASGFTR MQVALSVMKT AIPLVLLTMA IGEWVAPQGE QMARNYRAQA MYGGSLLSTQ QGLWAKDGNN F VYIERVKG DEVLGGISIY AFNENRRLQS VRYAATAKFD PEHKVWRLSQ VDESDLTNPK QITGSQTVSG TWKTDLTPDK LG VVALDPD ALSISGLHNY VKYLKSSGQD AGRYQLNMWS KIFQPLSVAV MMLMALSFIF GPLRSVPMGV RVVTGISFGF VFY VLDQIF GPLTLVYGIP PIIGALLPSA SFFLISLWLL MRKS UniProtKB: Inner membrane protein yjgQ |
-Macromolecule #4: PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER
| Macromolecule | Name: PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER / type: ligand / ID: 4 / Number of copies: 2 / Formula: ANP |
|---|---|
| Molecular weight | Theoretical: 506.196 Da |
| Chemical component information |  ChemComp-ANP: |
-Macromolecule #5: tetradecyl 4-O-alpha-D-glucopyranosyl-beta-D-glucopyranoside
| Macromolecule | Name: tetradecyl 4-O-alpha-D-glucopyranosyl-beta-D-glucopyranoside type: ligand / ID: 5 / Number of copies: 1 / Formula: LMD |
|---|---|
| Molecular weight | Theoretical: 538.669 Da |
| Chemical component information | 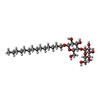 ChemComp-LMD: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1 mg/mL | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.8 Component:
| ||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 95 % / Chamber temperature: 298 K / Instrument: FEI VITROBOT MARK III |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Average electron dose: 40.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: DIFFRACTION |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: PDB ENTRY PDB model - PDB ID: |
|---|---|
| Final reconstruction | Applied symmetry - Point group: C1 (asymmetric) / Resolution.type: BY AUTHOR / Resolution: 3.5 Å / Resolution method: FSC 0.143 CUT-OFF / Software - Name: RELION (ver. 3) / Number images used: 149178 |
| Initial angle assignment | Type: ANGULAR RECONSTITUTION |
| Final angle assignment | Type: ANGULAR RECONSTITUTION |
 Movie
Movie Controller
Controller


















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)






















