[English] 日本語
 Yorodumi
Yorodumi- EMDB-1009: A cryo-electron microscopic study of ribosome-bound termination f... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-1009 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | A cryo-electron microscopic study of ribosome-bound termination factor RF2. | |||||||||
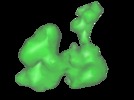
 Map data Map data | The RF2(GAQ) mutant cryo-density from the 3D-EM map of RF2 mutant bound to the E.coli 70s ribosome | |||||||||
 Sample Sample |
| |||||||||
| Function / homology |  Function and homology information Function and homology informationtranslation release factor activity, codon specific / translational termination / small ribosomal subunit / large ribosomal subunit rRNA binding / cytosolic large ribosomal subunit / tRNA binding / rRNA binding / structural constituent of ribosome / translation / viral translational frameshifting / cytosol Similarity search - Function | |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 10.9 Å | |||||||||
 Authors Authors | Rawat UB / Zavialov AV / Sengupta J / Valle M / Grassucci RA / Linde J / Vestergaard B / Ehrenberg M / Frank J | |||||||||
 Citation Citation |  Journal: Nature / Year: 2003 Journal: Nature / Year: 2003Title: A cryo-electron microscopic study of ribosome-bound termination factor RF2. Authors: Urmila B S Rawat / Andrey V Zavialov / Jayati Sengupta / Mikel Valle / Robert A Grassucci / Jamie Linde / Bente Vestergaard / Måns Ehrenberg / Joachim Frank /  Abstract: Protein synthesis takes place on the ribosome, where genetic information carried by messenger RNA is translated into a sequence of amino acids. This process is terminated when a stop codon moves into ...Protein synthesis takes place on the ribosome, where genetic information carried by messenger RNA is translated into a sequence of amino acids. This process is terminated when a stop codon moves into the ribosomal decoding centre (DC) and is recognized by a class-1 release factor (RF). RFs have a conserved GGQ amino-acid motif, which is crucial for peptide release and is believed to interact directly with the peptidyl-transferase centre (PTC) of the 50S ribosomal subunit. Another conserved motif of RFs (SPF in RF2) has been proposed to interact directly with stop codons in the DC of the 30S subunit. The distance between the DC and PTC is approximately 73 A. However, in the X-ray structure of RF2, SPF and GGQ are only 23 A apart, indicating that they cannot be at DC and PTC simultaneously. Here we show that RF2 is in an open conformation when bound to the ribosome, allowing GGQ to reach the PTC while still allowing SPF-stop-codon interaction. The results indicate new interpretations of accuracy in termination, and have implications for how the presence of a stop codon in the DC is signalled to PTC. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_1009.map.gz emd_1009.map.gz | 7.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-1009-v30.xml emd-1009-v30.xml emd-1009.xml emd-1009.xml | 8.8 KB 8.8 KB | Display Display |  EMDB header EMDB header |
| Images |  1009.gif 1009.gif | 15.3 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-1009 http://ftp.pdbj.org/pub/emdb/structures/EMD-1009 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-1009 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-1009 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_1009.map.gz / Format: CCP4 / Size: 8.2 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_1009.map.gz / Format: CCP4 / Size: 8.2 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | The RF2(GAQ) mutant cryo-density from the 3D-EM map of RF2 mutant bound to the E.coli 70s ribosome | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2.82 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : RF2 (GAQ) mutant from E.coli
| Entire | Name: RF2 (GAQ) mutant from E.coli |
|---|---|
| Components |
|
-Supramolecule #1000: RF2 (GAQ) mutant from E.coli
| Supramolecule | Name: RF2 (GAQ) mutant from E.coli / type: sample / ID: 1000 Details: SPIDER was used for the computational isolation of the RF2(GAQ) mutant cryo-density from the 3D-EM map of RF2 mutant bound to the E.coli 70s ribosome (EMD-1008). Number unique components: 1 |
|---|
-Macromolecule #1: RF2 (GAQ) mutant
| Macromolecule | Name: RF2 (GAQ) mutant / type: protein_or_peptide / ID: 1 / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:  |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Vitrification | Cryogen name: ETHANE |
|---|
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI F20 |
|---|---|
| Temperature | Average: 93 K |
| Details | information is the same as in submission EMD-1008 |
| Date | Aug 2, 2001 |
| Image recording | Category: FILM / Film or detector model: KODAK SO-163 FILM / Digitization - Scanner: ZEISS SCAI / Average electron dose: 20 e/Å2 / Details: entry EMD-1008 |
| Tilt angle min | 0 |
| Tilt angle max | 0 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated magnification: 49696 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.0 mm / Nominal defocus max: 4.05 µm / Nominal defocus min: 2.25 µm / Nominal magnification: 50000 |
| Sample stage | Specimen holder: Oxford, cryo-transfer 3500 / Specimen holder model: GATAN LIQUID NITROGEN |
| Experimental equipment |  Model: Tecnai F20 / Image courtesy: FEI Company |
- Image processing
Image processing
| Details | entry EMD-1008 |
|---|---|
| Final reconstruction | Applied symmetry - Point group: C1 (asymmetric) / Resolution.type: BY AUTHOR / Resolution: 10.9 Å Details: information is the same as in submission 1186. SPIDER was used for the computational isolation of the RF2 (GAQ) mutant cryo-density from the 3D-EM map of RF2 mutant bound to the E.coli 70s ribosome (EMD-1008). Number images used: 1 |
-Atomic model buiding 1
| Initial model | (PDB ID: , ) |
|---|---|
| Output model |  PDB-1mi6:  PDB-1mvr: |
 Movie
Movie Controller
Controller





















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)






















