[English] 日本語
 Yorodumi
Yorodumi- EMDB-0917: Cryo-EM structure of the human glucagon receptor in complex with Gs -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-0917 | |||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of the human glucagon receptor in complex with Gs | |||||||||||||||||||||||||||
 Map data Map data | ||||||||||||||||||||||||||||
 Sample Sample |
| |||||||||||||||||||||||||||
 Keywords Keywords | glucagon receptor / GPCR / Gs protein / SIGNALING PROTEIN | |||||||||||||||||||||||||||
| Function / homology |  Function and homology information Function and homology informationglucagon receptor binding / regulation of glycogen metabolic process / glucagon receptor activity / negative regulation of execution phase of apoptosis / feeding behavior / response to starvation / positive regulation of calcium ion import / positive regulation of insulin secretion involved in cellular response to glucose stimulus / exocytosis / PKA activation in glucagon signalling ...glucagon receptor binding / regulation of glycogen metabolic process / glucagon receptor activity / negative regulation of execution phase of apoptosis / feeding behavior / response to starvation / positive regulation of calcium ion import / positive regulation of insulin secretion involved in cellular response to glucose stimulus / exocytosis / PKA activation in glucagon signalling / peptide hormone binding / hair follicle placode formation / developmental growth / D1 dopamine receptor binding / Synthesis, secretion, and deacylation of Ghrelin / intracellular transport / renal water homeostasis / Hedgehog 'off' state / adenylate cyclase-activating adrenergic receptor signaling pathway / activation of adenylate cyclase activity / positive regulation of gluconeogenesis / cellular response to glucagon stimulus / protein kinase A signaling / adenylate cyclase activator activity / hormone-mediated signaling pathway / cellular response to starvation / regulation of insulin secretion / response to nutrient / trans-Golgi network membrane / guanyl-nucleotide exchange factor activity / positive regulation of peptidyl-threonine phosphorylation / generation of precursor metabolites and energy / response to activity / gluconeogenesis / negative regulation of inflammatory response to antigenic stimulus / bone development / G-protein beta/gamma-subunit complex binding / Olfactory Signaling Pathway / adenylate cyclase-modulating G protein-coupled receptor signaling pathway / Activation of the phototransduction cascade / adenylate cyclase-activating G protein-coupled receptor signaling pathway / hormone activity / G beta:gamma signalling through PLC beta / Presynaptic function of Kainate receptors / Thromboxane signalling through TP receptor / G protein-coupled acetylcholine receptor signaling pathway / G protein activity / G-protein activation / platelet aggregation / Activation of G protein gated Potassium channels / Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits / Prostacyclin signalling through prostacyclin receptor / Glucagon signaling in metabolic regulation / G beta:gamma signalling through CDC42 / regulation of blood pressure / cognition / G beta:gamma signalling through BTK / ADP signalling through P2Y purinoceptor 12 / Sensory perception of sweet, bitter, and umami (glutamate) taste / Synthesis, secretion, and inactivation of Glucagon-like Peptide-1 (GLP-1) / photoreceptor disc membrane / Glucagon-type ligand receptors / Adrenaline,noradrenaline inhibits insulin secretion / Vasopressin regulates renal water homeostasis via Aquaporins / G alpha (z) signalling events / Glucagon-like Peptide-1 (GLP1) regulates insulin secretion / cellular response to catecholamine stimulus / ADORA2B mediated anti-inflammatory cytokines production / sensory perception of taste / ADP signalling through P2Y purinoceptor 1 / G beta:gamma signalling through PI3Kgamma / adenylate cyclase-activating dopamine receptor signaling pathway / Cooperation of PDCL (PhLP1) and TRiC/CCT in G-protein beta folding / GPER1 signaling / cellular response to prostaglandin E stimulus / Inactivation, recovery and regulation of the phototransduction cascade / G-protein beta-subunit binding / heterotrimeric G-protein complex / sensory perception of smell / G alpha (12/13) signalling events / extracellular vesicle / signaling receptor complex adaptor activity / Thrombin signalling through proteinase activated receptors (PARs) / positive regulation of peptidyl-serine phosphorylation / GTPase binding / positive regulation of cold-induced thermogenesis / glucose homeostasis / retina development in camera-type eye / Ca2+ pathway / phospholipase C-activating G protein-coupled receptor signaling pathway / G alpha (i) signalling events / fibroblast proliferation / G alpha (s) signalling events / Hydrolases; Acting on acid anhydrides; Acting on GTP to facilitate cellular and subcellular movement / G alpha (q) signalling events / secretory granule lumen / Ras protein signal transduction / cell population proliferation / Extra-nuclear estrogen signaling / positive regulation of ERK1 and ERK2 cascade Similarity search - Function | |||||||||||||||||||||||||||
| Biological species |  Homo sapiens (human) / Homo sapiens (human) /  | |||||||||||||||||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.7 Å | |||||||||||||||||||||||||||
 Authors Authors | Qiao A / Han S | |||||||||||||||||||||||||||
| Funding support |  China, 8 items China, 8 items
| |||||||||||||||||||||||||||
 Citation Citation |  Journal: Science / Year: 2020 Journal: Science / Year: 2020Title: Structural basis of G and G recognition by the human glucagon receptor. Authors: Anna Qiao / Shuo Han / Xinmei Li / Zhixin Li / Peishen Zhao / Antao Dai / Rulve Chang / Linhua Tai / Qiuxiang Tan / Xiaojing Chu / Limin Ma / Thor Seneca Thorsen / Steffen Reedtz-Runge / ...Authors: Anna Qiao / Shuo Han / Xinmei Li / Zhixin Li / Peishen Zhao / Antao Dai / Rulve Chang / Linhua Tai / Qiuxiang Tan / Xiaojing Chu / Limin Ma / Thor Seneca Thorsen / Steffen Reedtz-Runge / Dehua Yang / Ming-Wei Wang / Patrick M Sexton / Denise Wootten / Fei Sun / Qiang Zhao / Beili Wu /    Abstract: Class B G protein-coupled receptors, an important class of therapeutic targets, signal mainly through the G class of heterotrimeric G proteins, although they do display some promiscuity in G protein ...Class B G protein-coupled receptors, an important class of therapeutic targets, signal mainly through the G class of heterotrimeric G proteins, although they do display some promiscuity in G protein binding. Using cryo-electron microscopy, we determined the structures of the human glucagon receptor (GCGR) bound to glucagon and distinct classes of heterotrimeric G proteins, G or G These two structures adopt a similar open binding cavity to accommodate G and G The G binding selectivity of GCGR is explained by a larger interaction interface, but there are specific interactions that affect G more than G binding. Conformational differences in the receptor intracellular loops were found to be key selectivity determinants. These distinctions in transducer engagement were supported by mutagenesis and functional studies. | |||||||||||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_0917.map.gz emd_0917.map.gz | 58.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-0917-v30.xml emd-0917-v30.xml emd-0917.xml emd-0917.xml | 18.3 KB 18.3 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_0917.png emd_0917.png | 45.5 KB | ||
| Masks |  emd_0917_msk_1.map emd_0917_msk_1.map | 64 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-0917.cif.gz emd-0917.cif.gz | 6.6 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-0917 http://ftp.pdbj.org/pub/emdb/structures/EMD-0917 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-0917 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-0917 | HTTPS FTP |
-Validation report
| Summary document |  emd_0917_validation.pdf.gz emd_0917_validation.pdf.gz | 559.3 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_0917_full_validation.pdf.gz emd_0917_full_validation.pdf.gz | 558.9 KB | Display | |
| Data in XML |  emd_0917_validation.xml.gz emd_0917_validation.xml.gz | 5.9 KB | Display | |
| Data in CIF |  emd_0917_validation.cif.gz emd_0917_validation.cif.gz | 6.7 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-0917 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-0917 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-0917 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-0917 | HTTPS FTP |
-Related structure data
| Related structure data |  6lmkMC  0918C  6lmlC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_0917.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_0917.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.82 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
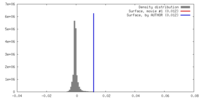
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_0917_msk_1.map emd_0917_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
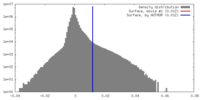
| Density Histograms |
- Sample components
Sample components
-Entire : Complex of glucagon receptor bound to glucagon, Gs protein and na...
| Entire | Name: Complex of glucagon receptor bound to glucagon, Gs protein and nanobody |
|---|---|
| Components |
|
-Supramolecule #1: Complex of glucagon receptor bound to glucagon, Gs protein and na...
| Supramolecule | Name: Complex of glucagon receptor bound to glucagon, Gs protein and nanobody type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Guanine nucleotide-binding protein G(s) subunit alpha isoforms short
| Macromolecule | Name: Guanine nucleotide-binding protein G(s) subunit alpha isoforms short type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 45.683434 KDa |
| Recombinant expression | Organism:  unidentified baculovirus unidentified baculovirus |
| Sequence | String: MGCLGNSKTE DQRNEEKAQR EANKKIEKQL QKDKQVYRAT HRLLLLGAGE SGKNTIVKQM RILHVNGFNG EGGEEDPQAA RSNSDGEKA TKVQDIKNNL KEAIETIVAA MSNLVPPVEL ANPENQFRVD YILSVMNVPD FDFPPEFYEH AKALWEDEGV R ACYERSNE ...String: MGCLGNSKTE DQRNEEKAQR EANKKIEKQL QKDKQVYRAT HRLLLLGAGE SGKNTIVKQM RILHVNGFNG EGGEEDPQAA RSNSDGEKA TKVQDIKNNL KEAIETIVAA MSNLVPPVEL ANPENQFRVD YILSVMNVPD FDFPPEFYEH AKALWEDEGV R ACYERSNE YQLIDCAQYF LDKIDVIKQA DYVPSDQDLL RCRVLTSGIF ETKFQVDKVN FHMFDVGAQR DERRKWIQCF ND VTAIIFV VASSSYNMVI REDNQTNRLQ AALKLFDSIW NNKWLRDTSV ILFLNKQDLL AEKVLAGKSK IEDYFPEFAR YTT PEDATP EPGEDPRVTR AKYFIRDEFL RISTASGDGR HYCYPHFTCA VDTENIRRVF NDCRDIIQRM HLRQYELL UniProtKB: Guanine nucleotide-binding protein G(s) subunit alpha isoforms short |
-Macromolecule #2: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1
| Macromolecule | Name: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 38.744371 KDa |
| Recombinant expression | Organism:  unidentified baculovirus unidentified baculovirus |
| Sequence | String: MHHHHHHGSL LQSELDQLRQ EAEQLKNQIR DARKACADAT LSQITNNIDP VGRIQMRTRR TLRGHLAKIY AMHWGTDSRL LVSASQDGK LIIWDSYTTN KVHAIPLRSS WVMTCAYAPS GNYVACGGLD NICSIYNLKT REGNVRVSRE LAGHTGYLSC C RFLDDNQI ...String: MHHHHHHGSL LQSELDQLRQ EAEQLKNQIR DARKACADAT LSQITNNIDP VGRIQMRTRR TLRGHLAKIY AMHWGTDSRL LVSASQDGK LIIWDSYTTN KVHAIPLRSS WVMTCAYAPS GNYVACGGLD NICSIYNLKT REGNVRVSRE LAGHTGYLSC C RFLDDNQI VTSSGDTTCA LWDIETGQQT TTFTGHTGDV MSLSLAPDTR LFVSGACDAS AKLWDVREGM CRQTFTGHES DI NAICFFP NGNAFATGSD DATCRLFDLR ADQELMTYSH DNIICGITSV SFSKSGRLLL AGYDDFNCNV WDALKADRAG VLA GHDNRV SCLGVTDDGM AVATGSWDSF LKIWN UniProtKB: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 |
-Macromolecule #3: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2
| Macromolecule | Name: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2 type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 7.861143 KDa |
| Recombinant expression | Organism:  unidentified baculovirus unidentified baculovirus |
| Sequence | String: MASNNTASIA QARKLVEQLK MEANIDRIKV SKAAADLMAY CEAHAKEDPL LTPVPASENP FREKKFFCAI L UniProtKB: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2 |
-Macromolecule #4: Nb35
| Macromolecule | Name: Nb35 / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 15.140742 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: QVQLQESGGG LVQPGGSLRL SCAASGFTFS NYKMNWVRQA PGKGLEWVSD ISQSGASISY TGSVKGRFTI SRDNAKNTLY LQMNSLKPE DTAVYYCARC PAPFTRDCFD VTSTTYAYRG QGTQVTVSSH HHHHHEPEA |
-Macromolecule #5: Glucagon receptor
| Macromolecule | Name: Glucagon receptor / type: protein_or_peptide / ID: 5 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 48.398109 KDa |
| Recombinant expression | Organism:  unidentified baculovirus unidentified baculovirus |
| Sequence | String: QVMDFLFEKW KLYGDQCHHN LSLLPPPTEL VCNRTFDKYS CWPDTPANTT ANISCPWYLP WHHKVQHRFV FKRCGPDGQW VRGPRGQPW RDASQCQMDG EEIEVQKEVA KMYSSFQVMY TVGYSLSLGA LLLALAILGG LSKLHCTRNA IHANLFASFV L KASSVLVI ...String: QVMDFLFEKW KLYGDQCHHN LSLLPPPTEL VCNRTFDKYS CWPDTPANTT ANISCPWYLP WHHKVQHRFV FKRCGPDGQW VRGPRGQPW RDASQCQMDG EEIEVQKEVA KMYSSFQVMY TVGYSLSLGA LLLALAILGG LSKLHCTRNA IHANLFASFV L KASSVLVI DGLLRTRYSQ KIGDDLSVST WLSDGAVAGC RVAAVFMQYG IVANYCWLLV EGLYLHNLLG LATLPERSFF SL YLGIGWG APMLFVVPWA VVKCLFENVQ CWTSNDNMGF WWILRFPVFL AILINFFIFV RIVQLLVAKL RARQMHHTDY KFR LAKSTL TLIPLLGVHE VVFAFVTDEH AQGTLRSAKL FFDLFLSSFQ GLLVAVLYCF LNKEVQSELR RRWHRWRLGK VLWE ERNTS NGSGSEDQVD PRLIDGK UniProtKB: Glucagon receptor |
-Macromolecule #6: Glucagon
| Macromolecule | Name: Glucagon / type: protein_or_peptide / ID: 6 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 3.486781 KDa |
| Sequence | String: HSQGTFTSDY SKYLDSRRAQ DFVQWLMNT UniProtKB: Pro-glucagon |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 1.875 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: PDB ENTRY PDB model - PDB ID: |
|---|---|
| Final reconstruction | Applied symmetry - Point group: C1 (asymmetric) / Resolution.type: BY AUTHOR / Resolution: 3.7 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 169878 |
| Initial angle assignment | Type: ANGULAR RECONSTITUTION |
| Final angle assignment | Type: ANGULAR RECONSTITUTION |
 Movie
Movie Controller
Controller





































 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)