+Search query
-Structure paper
| Title | Design of potent selective zinc-mediated serine protease inhibitors. |
|---|---|
| Journal, issue, pages | Nature, Vol. 391, Page 608-612, Year 1998 |
| Publish date | Jul 21, 1999 (structure data deposition date) |
 Authors Authors | Katz, B.A. / Clark, J.M. / Finer-Moore, J.S. / Jenkins, T.E. / Johnson, C.R. / Ross, M.J. / Luong, C. / Moore, W.R. / Stroud, R.M. |
 External links External links |  Nature / Nature /  PubMed:9468142 PubMed:9468142 |
| Methods | X-ray diffraction |
| Resolution | 1.37 - 2.2 Å |
| Structure data |  PDB-1c1n:  PDB-1c1o:  PDB-1c1p:  PDB-1c1q:  PDB-1c1r:  PDB-1c1t:  PDB-1c1u:  PDB-1c1v:  PDB-1c1w:  PDB-1c2d:  PDB-1c2e:  PDB-1c2f:  PDB-1c2g:  PDB-1c2h:  PDB-1c2i:  PDB-1c2j:  PDB-1c2k:  PDB-1c2l:  PDB-1c2m:  PDB-1xuf:  PDB-1xug:  PDB-1xuh:  PDB-1xui:  PDB-1xuj:  PDB-1xuk: |
| Chemicals |  ChemComp-CA:  ChemComp-ZN:  ChemComp-SO4:  ChemComp-BEN:  ChemComp-HOH:  ChemComp-MG:  ChemComp-BAI:  ChemComp-DMS:  ChemComp-BAB:  ChemComp-NA:  ChemComp-BAH:  ChemComp-BAK:  ChemComp-CL:  ChemComp-ABI: 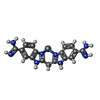 ChemComp-BAZ:  ChemComp-CO: 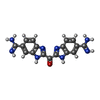 ChemComp-BAO:  ChemComp-BOZ: |
| Source |
|
 Keywords Keywords | HYDROLASE/HYDROLASE INHIBITOR / ZN(II)-MEDIATED SERINE PROTEASE INHIBITORS / PH DEPENDENCE / ZN(II) AFFINITY STUCTURE-BASED DRUG DESIGN / SERINE PROTEASE / HYDROLASE-HYDROLASE INHIBITOR COMPLEX / SERINE PROTEASE SERINE PROTEASE/INHIBITOR / SERINE PROTEASE/INHIBITOR / BLOOD CLOTTING/HYDROLASE INHIBITOR / BLOOD CLOTTING-HYDROLASE INHIBITOR COMPLEX / BLOOD CLOTTING / COMPLEX / TRYPSIN-ZN+2-SMALL MOLECULE LIGAND / DESIGNED SMALL MOLECULE LIGAND WITH NANOMOLAR AFFINITY / TRYPSIN-COBALT-SMALL MOLECULE LIGAND / DESIGNED SMALL MOLECULE LIGAND WITH MICROMOLAR AFFINITY / TRYPSIN-SMALL MOLECULE LIGAND / TRYPSIN-SULFATE-SMALL MOLECULE LIGAND |
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About Yorodumi Papers
About Yorodumi Papers




 homo sapiens (human)
homo sapiens (human) hirudo medicinalis (medicinal leech)
hirudo medicinalis (medicinal leech)