+Search query
-Structure paper
| Title | Structural insights into dimerization and activation of the mGlu2-mGlu3 and mGlu2-mGlu4 heterodimers. |
|---|---|
| Journal, issue, pages | Cell Res, Vol. 33, Issue 10, Page 762-774, Year 2023 |
| Publish date | Jun 8, 2023 |
 Authors Authors | Xinwei Wang / Mu Wang / Tuo Xu / Ye Feng / Qiang Shao / Shuo Han / Xiaojing Chu / Yechun Xu / Shuling Lin / Qiang Zhao / Beili Wu /  |
| PubMed Abstract | Heterodimerization of the metabotropic glutamate receptors (mGlus) has shown importance in the functional modulation of the receptors and offers potential drug targets for treating central nervous ...Heterodimerization of the metabotropic glutamate receptors (mGlus) has shown importance in the functional modulation of the receptors and offers potential drug targets for treating central nervous system diseases. However, due to a lack of molecular details of the mGlu heterodimers, understanding of the mechanisms underlying mGlu heterodimerization and activation is limited. Here we report twelve cryo-electron microscopy (cryo-EM) structures of the mGlu2-mGlu3 and mGlu2-mGlu4 heterodimers in different conformational states, including inactive, intermediate inactive, intermediate active and fully active conformations. These structures provide a full picture of conformational rearrangement of mGlu2-mGlu3 upon activation. The Venus flytrap domains undergo a sequential conformational change, while the transmembrane domains exhibit a substantial rearrangement from an inactive, symmetric dimer with diverse dimerization patterns to an active, asymmetric dimer in a conserved dimerization mode. Combined with functional data, these structures reveal that stability of the inactive conformations of the subunits and the subunit-G protein interaction pattern are determinants of asymmetric signal transduction of the heterodimers. Furthermore, a novel binding site for two mGlu4 positive allosteric modulators was observed in the asymmetric dimer interfaces of the mGlu2-mGlu4 heterodimer and mGlu4 homodimer, and may serve as a drug recognition site. These findings greatly extend our knowledge about signal transduction of the mGlus. |
 External links External links |  Cell Res / Cell Res /  PubMed:37286794 / PubMed:37286794 /  PubMed Central PubMed Central |
| Methods | EM (single particle) |
| Resolution | 2.8 - 3.7 Å |
| Structure data | EMDB-36165, PDB-8jcu: EMDB-36166, PDB-8jcv: EMDB-36167, PDB-8jcw: EMDB-36168, PDB-8jcx: EMDB-36169, PDB-8jcy: EMDB-36170, PDB-8jcz: EMDB-36171, PDB-8jd0: EMDB-36172, PDB-8jd1: EMDB-36173, PDB-8jd2: EMDB-36174, PDB-8jd3: EMDB-36175, PDB-8jd4: EMDB-36176, PDB-8jd5: EMDB-36177, PDB-8jd6:  EMDB-36221: Cryo-EM map of Gi-bound metabotropic glutamate receptor mGlu4 focused on TMD and G protein  EMDB-36222: Cryo-EM map of Gi1-bound metabotropic glutamate receptor mGlu4 focused on ECD  EMDB-36226: Cryo-EM map of Gi1-bound mGlu2-mGlu4 heterodimer focused on ECD  EMDB-36227: Cryo-EM map of Gi1-bound mGlu2-mGlu4 heterodimer focused on TMD and G protein  EMDB-36286: The consensus Cryo-EM map of Gi1-bound mGlu2-mGlu4 heterodimer  EMDB-36287: The consensus Cryo-EM map of Gi-bound metabotropic glutamate receptor mGlu4 |
| Chemicals | 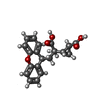 ChemComp-Z99:  ChemComp-NAG:  ChemComp-CLR: 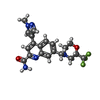 ChemComp-J9R:  ChemComp-GLU:  ChemComp-HZR:  ChemComp-PEF:  ChemComp-BQI:  ChemComp-BK0: 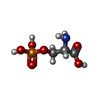 ChemComp-SEP: |
| Source |
|
 Keywords Keywords | MEMBRANE PROTEIN / Complex structure / mGlu2-3 heterodimer / mGlu2-3 heterodimer in presence of NAM563 / mGlu2-mGlu3 heterodimer with Gi protein / mGlu2-mGlu4 heterodimer / Gi1-bound mGlu2-mGlu4 heterodimer / Gi-bound metabotropic glutamate receptor mGlu4 |
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About Yorodumi Papers
About Yorodumi Papers





























 homo sapiens (human)
homo sapiens (human)
