+Search query
-Structure paper
| Title | Structure and activation of the RING E3 ubiquitin ligase TRIM72 on the membrane. |
|---|---|
| Journal, issue, pages | Nat Struct Mol Biol, Vol. 30, Issue 11, Page 1695-1706, Year 2023 |
| Publish date | Sep 28, 2023 |
 Authors Authors | Si Hoon Park / Juhyun Han / Byung-Cheon Jeong / Ju Han Song / Se Hwan Jang / Hyeongseop Jeong / Bong Heon Kim / Young-Gyu Ko / Zee-Yong Park / Kyung Eun Lee / Jaekyung Hyun / Hyun Kyu Song /    |
| PubMed Abstract | Defects in plasma membrane repair can lead to muscle and heart diseases in humans. Tripartite motif-containing protein (TRIM)72 (mitsugumin 53; MG53) has been determined to rapidly nucleate vesicles ...Defects in plasma membrane repair can lead to muscle and heart diseases in humans. Tripartite motif-containing protein (TRIM)72 (mitsugumin 53; MG53) has been determined to rapidly nucleate vesicles at the site of membrane damage, but the underlying molecular mechanisms remain poorly understood. Here we present the structure of Mus musculus TRIM72, a complete model of a TRIM E3 ubiquitin ligase. We demonstrated that the interaction between TRIM72 and phosphatidylserine-enriched membranes is necessary for its oligomeric assembly and ubiquitination activity. Using cryogenic electron tomography and subtomogram averaging, we elucidated a higher-order model of TRIM72 assembly on the phospholipid bilayer. Combining structural and biochemical techniques, we developed a working molecular model of TRIM72, providing insights into the regulation of RING-type E3 ligases through the cooperation of multiple domains in higher-order assemblies. Our findings establish a fundamental basis for the study of TRIM E3 ligases and have therapeutic implications for diseases associated with membrane repair. |
 External links External links |  Nat Struct Mol Biol / Nat Struct Mol Biol /  PubMed:37770719 / PubMed:37770719 /  PubMed Central PubMed Central |
| Methods | EM (tomography) / EM (subtomogram averaging) / X-ray diffraction |
| Resolution | 2.75 - 26.0 Å |
| Structure data |  EMDB-31139: Reconstituted proteoliposomes of TRIM72 in negative curvature #1 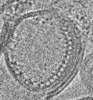 EMDB-31150: Reconstituted proteoliposomes of TRIM72 in negative curvature #2 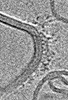 EMDB-31151: Reconstituted proteoliposomes of TRIM72 in positive curvature #1 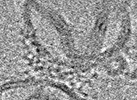 EMDB-31152: Reconstituted proteoliposomes of TRIM72 in positive curvature #2  EMDB-33569: Higher-ordered assembly of mouse TRIM72 WT on the Phosphatidylserine/Cholesterol liposome bilayer  EMDB-33582: Higher-ordered assembly of mouse TRIM72 M138R on the Phosphatidylserine/Cholesterol liposome bilayer  PDB-7xv2:  PDB-7xyy:  PDB-7xyz:  PDB-7xz0:  PDB-7xz1:  PDB-7xz2: |
| Chemicals |  ChemComp-ZN: |
| Source |
|
 Keywords Keywords | MEMBRANE PROTEIN / TRIM / Tripartite motif / Ubiquitin ligase / Coiled coil / B-box / PRY-SPRY / LIGASE / METAL BINDING PROTEIN / TRIM72 / MG53 |
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About Yorodumi Papers
About Yorodumi Papers




