[English] 日本語
 Yorodumi
Yorodumi- EMDB-33582: Higher-ordered assembly of mouse TRIM72 M138R on the Phosphatidyl... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Higher-ordered assembly of mouse TRIM72 M138R on the Phosphatidylserine/Cholesterol liposome bilayer | |||||||||
 Map data Map data | EM map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | TRIM / Tripartite motif / Ubiquitin ligase / Coiled coil / B-box / PRY-SPRY / Membrane protein / LIGASE / METAL BINDING PROTEIN / Phosphatidylserine / TRIM72 / MG53 | |||||||||
| Biological species |  | |||||||||
| Method | subtomogram averaging / cryo EM / Resolution: 26.0 Å | |||||||||
 Authors Authors | Park SH / Hyun J / Jeong H / Song HK | |||||||||
| Funding support |  Korea, Republic Of, 1 items Korea, Republic Of, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Struct Mol Biol / Year: 2023 Journal: Nat Struct Mol Biol / Year: 2023Title: Structure and activation of the RING E3 ubiquitin ligase TRIM72 on the membrane. Authors: Si Hoon Park / Juhyun Han / Byung-Cheon Jeong / Ju Han Song / Se Hwan Jang / Hyeongseop Jeong / Bong Heon Kim / Young-Gyu Ko / Zee-Yong Park / Kyung Eun Lee / Jaekyung Hyun / Hyun Kyu Song /    Abstract: Defects in plasma membrane repair can lead to muscle and heart diseases in humans. Tripartite motif-containing protein (TRIM)72 (mitsugumin 53; MG53) has been determined to rapidly nucleate vesicles ...Defects in plasma membrane repair can lead to muscle and heart diseases in humans. Tripartite motif-containing protein (TRIM)72 (mitsugumin 53; MG53) has been determined to rapidly nucleate vesicles at the site of membrane damage, but the underlying molecular mechanisms remain poorly understood. Here we present the structure of Mus musculus TRIM72, a complete model of a TRIM E3 ubiquitin ligase. We demonstrated that the interaction between TRIM72 and phosphatidylserine-enriched membranes is necessary for its oligomeric assembly and ubiquitination activity. Using cryogenic electron tomography and subtomogram averaging, we elucidated a higher-order model of TRIM72 assembly on the phospholipid bilayer. Combining structural and biochemical techniques, we developed a working molecular model of TRIM72, providing insights into the regulation of RING-type E3 ligases through the cooperation of multiple domains in higher-order assemblies. Our findings establish a fundamental basis for the study of TRIM E3 ligases and have therapeutic implications for diseases associated with membrane repair. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_33582.map.gz emd_33582.map.gz | 12.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-33582-v30.xml emd-33582-v30.xml emd-33582.xml emd-33582.xml | 15.1 KB 15.1 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_33582_fsc.xml emd_33582_fsc.xml | 6.9 KB | Display |  FSC data file FSC data file |
| Images |  emd_33582.png emd_33582.png | 33.7 KB | ||
| Masks |  emd_33582_msk_1.map emd_33582_msk_1.map | 27 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-33582.cif.gz emd-33582.cif.gz | 5.1 KB | ||
| Others |  emd_33582_half_map_1.map.gz emd_33582_half_map_1.map.gz emd_33582_half_map_2.map.gz emd_33582_half_map_2.map.gz | 20.6 MB 20.6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-33582 http://ftp.pdbj.org/pub/emdb/structures/EMD-33582 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-33582 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-33582 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_33582.map.gz / Format: CCP4 / Size: 27 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_33582.map.gz / Format: CCP4 / Size: 27 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | EM map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2.3 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_33582_msk_1.map emd_33582_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: half map 1
| File | emd_33582_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: half map 2
| File | emd_33582_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Higher-ordered assembly of mouse TRIM72 M138R on the Phosphatidyl...
| Entire | Name: Higher-ordered assembly of mouse TRIM72 M138R on the Phosphatidylserine/Cholesterol liposome bilayer |
|---|---|
| Components |
|
-Supramolecule #1: Higher-ordered assembly of mouse TRIM72 M138R on the Phosphatidyl...
| Supramolecule | Name: Higher-ordered assembly of mouse TRIM72 M138R on the Phosphatidylserine/Cholesterol liposome bilayer type: complex / ID: 1 / Parent: 0 / Macromolecule list: all Details: Higher-ordered assembly of mouse TRIM72 M138R on the Phosphatidylserine/Cholesterol liposome bilayer |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Mouse TRIM72 M138R
| Macromolecule | Name: Mouse TRIM72 M138R / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Recombinant expression | Organism:  |
| Sequence | String: GSAAPGLLRQ ELSCPLCLQL FDAPVTAECG HSFCRACLIR VAGEPAADGT VACPCCQAPT RPQALSTNLQ LSRLVEGLAQ VPQGHCEEH LDPLSIYCEQ DRTLVCGVCA SLGSHRGHRL LPAAEAQARL KTQLPQQKRQ LQEACMRKEK TVAVLEHQLV E VEETVRQF ...String: GSAAPGLLRQ ELSCPLCLQL FDAPVTAECG HSFCRACLIR VAGEPAADGT VACPCCQAPT RPQALSTNLQ LSRLVEGLAQ VPQGHCEEH LDPLSIYCEQ DRTLVCGVCA SLGSHRGHRL LPAAEAQARL KTQLPQQKRQ LQEACMRKEK TVAVLEHQLV E VEETVRQF RGAVGEQLGK MRMFLAALES SLDREAERVR GDAGVALRRE LSSLNSYLEQ LRQMEKVLEE VADKPQTEFL MK FCLVTSR LQKILSESPP PARLDIQLPV ISDDFKFQVW KKMFRALMPA LEELTFDPSS AHPSLVVSSS GRRVECSDQK APP AGEDTR QFDKAVAVVA QQLLSQGEHY WEVEVGDKPR WALGVMAADA SRRGRLHAVP SQGLWLLGLR DGKILEAHVE AKEP RALRT PERPPARIGL YLSFADGVLA FYDASNPDVL TPIFSFHERL PGPVYPIFDV CWHDKGKNAQ PLLLVGPEQE QA |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | subtomogram averaging |
| Aggregation state | 2D array |
- Sample preparation
Sample preparation
| Buffer | pH: 8 / Details: 50 mM Tris-HCl pH 8.0 150 mM NaCl 1 mM TCEP |
|---|---|
| Grid | Model: Quantifoil R1.2/1.3 / Support film - Material: CARBON / Support film - topology: HOLEY |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 95 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV / Details: blot for 7 seconds before plunging. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Average electron dose: 1.4 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 0.2 mm / Nominal defocus max: 6.0 µm / Nominal defocus min: 4.0 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: AB INITIO MODEL |
|---|
 Movie
Movie Controller
Controller




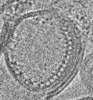
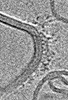
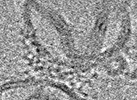







 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)