+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 7xz0 | ||||||
|---|---|---|---|---|---|---|---|
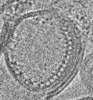
| Title | TRIM E3 ubiquitin ligase | ||||||
 Components Components | Tripartite motif-containing protein 72 | ||||||
 Keywords Keywords | MEMBRANE PROTEIN / TRIM / Tripartite motif / Ubiquitin ligase / Coiled coil / B-box / PRY-SPRY / LIGASE / METAL BINDING PROTEIN / TRIM72 / MG53 | ||||||
| Function / homology |  Function and homology information Function and homology informationmuscle system process / negative regulation of insulin-like growth factor receptor signaling pathway / negative regulation of myotube differentiation / plasma membrane repair / vesicle budding from membrane / ubiquitin conjugating enzyme binding / mitogen-activated protein kinase kinase kinase binding / muscle organ development / phosphatidylserine binding / exocytosis ...muscle system process / negative regulation of insulin-like growth factor receptor signaling pathway / negative regulation of myotube differentiation / plasma membrane repair / vesicle budding from membrane / ubiquitin conjugating enzyme binding / mitogen-activated protein kinase kinase kinase binding / muscle organ development / phosphatidylserine binding / exocytosis / negative regulation of insulin receptor signaling pathway / cytoplasmic vesicle membrane / protein homooligomerization / sarcolemma / RING-type E3 ubiquitin transferase / ubiquitin protein ligase activity / proteasome-mediated ubiquitin-dependent protein catabolic process / zinc ion binding / identical protein binding Similarity search - Function | ||||||
| Biological species |  | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 3.28 Å MOLECULAR REPLACEMENT / Resolution: 3.28 Å | ||||||
 Authors Authors | Park, S.H. / Song, H.K. | ||||||
| Funding support |  Korea, Republic Of, 1items Korea, Republic Of, 1items
| ||||||
 Citation Citation |  Journal: Nat Struct Mol Biol / Year: 2023 Journal: Nat Struct Mol Biol / Year: 2023Title: Structure and activation of the RING E3 ubiquitin ligase TRIM72 on the membrane. Authors: Si Hoon Park / Juhyun Han / Byung-Cheon Jeong / Ju Han Song / Se Hwan Jang / Hyeongseop Jeong / Bong Heon Kim / Young-Gyu Ko / Zee-Yong Park / Kyung Eun Lee / Jaekyung Hyun / Hyun Kyu Song /    Abstract: Defects in plasma membrane repair can lead to muscle and heart diseases in humans. Tripartite motif-containing protein (TRIM)72 (mitsugumin 53; MG53) has been determined to rapidly nucleate vesicles ...Defects in plasma membrane repair can lead to muscle and heart diseases in humans. Tripartite motif-containing protein (TRIM)72 (mitsugumin 53; MG53) has been determined to rapidly nucleate vesicles at the site of membrane damage, but the underlying molecular mechanisms remain poorly understood. Here we present the structure of Mus musculus TRIM72, a complete model of a TRIM E3 ubiquitin ligase. We demonstrated that the interaction between TRIM72 and phosphatidylserine-enriched membranes is necessary for its oligomeric assembly and ubiquitination activity. Using cryogenic electron tomography and subtomogram averaging, we elucidated a higher-order model of TRIM72 assembly on the phospholipid bilayer. Combining structural and biochemical techniques, we developed a working molecular model of TRIM72, providing insights into the regulation of RING-type E3 ligases through the cooperation of multiple domains in higher-order assemblies. Our findings establish a fundamental basis for the study of TRIM E3 ligases and have therapeutic implications for diseases associated with membrane repair. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  7xz0.cif.gz 7xz0.cif.gz | 388.9 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb7xz0.ent.gz pdb7xz0.ent.gz | 270.7 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  7xz0.json.gz 7xz0.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/xz/7xz0 https://data.pdbj.org/pub/pdb/validation_reports/xz/7xz0 ftp://data.pdbj.org/pub/pdb/validation_reports/xz/7xz0 ftp://data.pdbj.org/pub/pdb/validation_reports/xz/7xz0 | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  7xv2SC  7xyyC  7xyzC  7xz1C  7xz2C S: Starting model for refinement C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||||||||||||
| Unit cell |
| ||||||||||||||||||
| Noncrystallographic symmetry (NCS) | NCS domain:
NCS domain segments: Component-ID: 1 / Ens-ID: ens_1 / Beg auth comp-ID: HIS / Beg label comp-ID: HIS / End auth comp-ID: ARG / End label comp-ID: ARG / Auth seq-ID: 85 - 386 / Label seq-ID: 9 - 310
NCS oper: (Code: givenMatrix: (0.126394169972, 0.353672994327, -0.926790120189), (0.360635564223, -0.886736862431, -0.289205336435), (-0.924103080622, -0.297679609461, -0.23962542957)Vector: -7. ...NCS oper: (Code: given Matrix: (0.126394169972, 0.353672994327, -0.926790120189), Vector: |
- Components
Components
| #1: Protein | Mass: 44200.523 Da / Num. of mol.: 2 Source method: isolated from a genetically manipulated source Source: (gene. exp.)   #2: Chemical | ChemComp-ZN / Has ligand of interest | Y | |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 4.11 Å3/Da / Density % sol: 70.05 % |
|---|---|
| Crystal grow | Temperature: 277 K / Method: vapor diffusion, hanging drop / Details: 100 mM TRIS-HCl pH 8.5, 8% (v/v) PEG 8000 |
-Data collection
| Diffraction | Mean temperature: 100 K / Serial crystal experiment: N |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  Photon Factory Photon Factory  / Beamline: AR-NE3A / Wavelength: 1 Å / Beamline: AR-NE3A / Wavelength: 1 Å |
| Detector | Type: ADSC QUANTUM 315r / Detector: CCD / Date: May 30, 2014 |
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 1 Å / Relative weight: 1 |
| Reflection | Resolution: 3.28→50 Å / Num. obs: 22376 / % possible obs: 98.8 % / Redundancy: 3.9 % / Biso Wilson estimate: 100.76 Å2 / CC1/2: 0.995 / Net I/σ(I): 21.5 |
| Reflection shell | Resolution: 3.28→3.34 Å / Num. unique obs: 1110 / CC1/2: 0.558 |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: 7XV2 Resolution: 3.28→37.74 Å / SU ML: 0.5585 / Cross valid method: FREE R-VALUE / σ(F): 1.33 / Phase error: 36.2094 Stereochemistry target values: GeoStd + Monomer Library + CDL v1.2
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Shrinkage radii: 0.9 Å / VDW probe radii: 1.1 Å / Solvent model: FLAT BULK SOLVENT MODEL | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 139.02 Å2 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 3.28→37.74 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints NCS | Type: Torsion NCS / Rms dev position: 2.83355620019 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement TLS params. | Method: refined / Refine-ID: X-RAY DIFFRACTION
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement TLS group | Refine-ID: X-RAY DIFFRACTION
|
 Movie
Movie Controller
Controller






 PDBj
PDBj



