+Search query
-Structure paper
| Title | Distinct structure and gating mechanism in diverse NMDA receptors with GluN2C and GluN2D subunits. |
|---|---|
| Journal, issue, pages | Nat Struct Mol Biol, Vol. 30, Issue 5, Page 629-639, Year 2023 |
| Publish date | Mar 23, 2023 |
 Authors Authors | Jilin Zhang / Ming Zhang / Qinrui Wang / Han Wen / Zheyi Liu / Fangjun Wang / Yuhang Wang / Fenyong Yao / Nan Song / Zengwei Kou / Yang Li / Fei Guo / Shujia Zhu /  |
| PubMed Abstract | N-methyl-D-aspartate (NMDA) receptors are heterotetramers comprising two GluN1 and two alternate GluN2 (N2A-N2D) subunits. Here we report full-length cryo-EM structures of the human N1-N2D di- ...N-methyl-D-aspartate (NMDA) receptors are heterotetramers comprising two GluN1 and two alternate GluN2 (N2A-N2D) subunits. Here we report full-length cryo-EM structures of the human N1-N2D di-heterotetramer (di-receptor), rat N1-N2C di-receptor and N1-N2A-N2C tri-heterotetramer (tri-receptor) at a best resolution of 3.0 Å. The bilobate N-terminal domain (NTD) in N2D intrinsically adopts a closed conformation, leading to a compact NTD tetramer in the N1-N2D receptor. Additionally, crosslinking the ligand-binding domain (LBD) of two N1 protomers significantly elevated the channel open probability (Po) in N1-N2D di-receptors. Surprisingly, the N1-N2C di-receptor adopted both symmetric (minor) and asymmetric (major) conformations, the latter further locked by an allosteric potentiator, PYD-106, binding to a pocket between the NTD and LBD in only one N2C protomer. Finally, the N2A and N2C subunits in the N1-N2A-N2C tri-receptor display a conformation close to one protomer in the N1-N2A and N1-N2C di-receptors, respectively. These findings provide a comprehensive structural understanding of diverse function in major NMDA receptor subtypes. |
 External links External links |  Nat Struct Mol Biol / Nat Struct Mol Biol /  PubMed:36959261 PubMed:36959261 |
| Methods | EM (single particle) |
| Resolution | 3.0 - 6.4 Å |
| Structure data | EMDB-33788, PDB-7yff: EMDB-33789, PDB-7yfg: EMDB-33790, PDB-7yfh: EMDB-33791, PDB-7yfi: EMDB-33792, PDB-7yfl: EMDB-33793, PDB-7yfm: EMDB-33795, PDB-7yfo: EMDB-33798, PDB-7yfr: EMDB-34674, PDB-8hdk: |
| Chemicals |  ChemComp-NAG:  ChemComp-GLY: 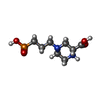 ChemComp-7RC:  ChemComp-GLU: 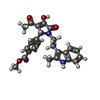 ChemComp-IWB: |
| Source |
|
 Keywords Keywords | ELECTRON TRANSPORT / ion channel / cryo-EM structure / glutamate receptor / synaptic protein / MEMBRANE PROTEIN / NMDA receptor / GluN2C / GluN2A |
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About Yorodumi Papers
About Yorodumi Papers





















 homo sapiens (human)
homo sapiens (human)
