[English] 日本語
 Yorodumi
Yorodumi- EMDB-33791: Structure of the Rat tri-heteromeric GluN1-GluN2A-GluN2C NMDA rec... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of the Rat tri-heteromeric GluN1-GluN2A-GluN2C NMDA receptor in complex with glycine and glutamate | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | NMDA receptor / GluN2C / GluN2A / MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationdirectional locomotion / positive regulation of Schwann cell migration / pons maturation / regulation of cell communication / EPHB-mediated forward signaling / Assembly and cell surface presentation of NMDA receptors / olfactory learning / conditioned taste aversion / dendritic branch / regulation of respiratory gaseous exchange ...directional locomotion / positive regulation of Schwann cell migration / pons maturation / regulation of cell communication / EPHB-mediated forward signaling / Assembly and cell surface presentation of NMDA receptors / olfactory learning / conditioned taste aversion / dendritic branch / regulation of respiratory gaseous exchange / protein localization to postsynaptic membrane / serotonin metabolic process / transmitter-gated monoatomic ion channel activity / suckling behavior / response to glycine / propylene metabolic process / sleep / RAF/MAP kinase cascade / regulation of monoatomic cation transmembrane transport / NMDA glutamate receptor activity / Synaptic adhesion-like molecules / voltage-gated monoatomic cation channel activity / response to glycoside / NMDA selective glutamate receptor complex / glutamate binding / ligand-gated sodium channel activity / neurotransmitter receptor complex / neuromuscular process controlling balance / response to morphine / regulation of axonogenesis / neuromuscular process / calcium ion transmembrane import into cytosol / regulation of dendrite morphogenesis / protein heterotetramerization / male mating behavior / regulation of synapse assembly / response to amine / glycine binding / startle response / parallel fiber to Purkinje cell synapse / positive regulation of reactive oxygen species biosynthetic process / dopamine metabolic process / monoatomic cation transmembrane transport / positive regulation of calcium ion transport into cytosol / cellular response to glycine / associative learning / excitatory synapse / positive regulation of dendritic spine maintenance / monoatomic ion channel complex / social behavior / monoatomic cation transport / regulation of neuronal synaptic plasticity / glutamate receptor binding / Unblocking of NMDA receptors, glutamate binding and activation / positive regulation of excitatory postsynaptic potential / long-term memory / synaptic cleft / phosphatase binding / prepulse inhibition / positive regulation of synaptic transmission, glutamatergic / monoatomic cation channel activity / response to fungicide / calcium ion homeostasis / glutamate-gated receptor activity / regulation of neuron apoptotic process / cellular response to manganese ion / glutamate-gated calcium ion channel activity / presynaptic active zone membrane / neurogenesis / dendrite membrane / sensory perception of pain / ligand-gated monoatomic ion channel activity involved in regulation of presynaptic membrane potential / ionotropic glutamate receptor signaling pathway / sodium ion transmembrane transport / synaptic membrane / response to amphetamine / cytoplasmic vesicle membrane / hippocampal mossy fiber to CA3 synapse / adult locomotory behavior / regulation of membrane potential / learning / PDZ domain binding / transmitter-gated monoatomic ion channel activity involved in regulation of postsynaptic membrane potential / synaptic transmission, glutamatergic / excitatory postsynaptic potential / protein catabolic process / response to calcium ion / regulation of long-term neuronal synaptic plasticity / postsynaptic density membrane / neuron cellular homeostasis / cerebral cortex development / regulation of synaptic plasticity / visual learning / negative regulation of protein catabolic process / calcium ion transmembrane transport / response to wounding / calcium channel activity / memory / long-term synaptic potentiation / terminal bouton Similarity search - Function | |||||||||
| Biological species |  | |||||||||
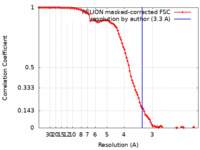
| Method | single particle reconstruction / cryo EM / Resolution: 3.3 Å | |||||||||
 Authors Authors | Zhang M / Zhang J / Guo F / Li Y / Zhu S | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Struct Mol Biol / Year: 2023 Journal: Nat Struct Mol Biol / Year: 2023Title: Distinct structure and gating mechanism in diverse NMDA receptors with GluN2C and GluN2D subunits. Authors: Jilin Zhang / Ming Zhang / Qinrui Wang / Han Wen / Zheyi Liu / Fangjun Wang / Yuhang Wang / Fenyong Yao / Nan Song / Zengwei Kou / Yang Li / Fei Guo / Shujia Zhu /  Abstract: N-methyl-D-aspartate (NMDA) receptors are heterotetramers comprising two GluN1 and two alternate GluN2 (N2A-N2D) subunits. Here we report full-length cryo-EM structures of the human N1-N2D di- ...N-methyl-D-aspartate (NMDA) receptors are heterotetramers comprising two GluN1 and two alternate GluN2 (N2A-N2D) subunits. Here we report full-length cryo-EM structures of the human N1-N2D di-heterotetramer (di-receptor), rat N1-N2C di-receptor and N1-N2A-N2C tri-heterotetramer (tri-receptor) at a best resolution of 3.0 Å. The bilobate N-terminal domain (NTD) in N2D intrinsically adopts a closed conformation, leading to a compact NTD tetramer in the N1-N2D receptor. Additionally, crosslinking the ligand-binding domain (LBD) of two N1 protomers significantly elevated the channel open probability (Po) in N1-N2D di-receptors. Surprisingly, the N1-N2C di-receptor adopted both symmetric (minor) and asymmetric (major) conformations, the latter further locked by an allosteric potentiator, PYD-106, binding to a pocket between the NTD and LBD in only one N2C protomer. Finally, the N2A and N2C subunits in the N1-N2A-N2C tri-receptor display a conformation close to one protomer in the N1-N2A and N1-N2C di-receptors, respectively. These findings provide a comprehensive structural understanding of diverse function in major NMDA receptor subtypes. #1:  Journal: Nat.Struct.Mol.Biol. / Year: 2023 Journal: Nat.Struct.Mol.Biol. / Year: 2023Title: Distinct structure and gating mechanism in diverse NMDA receptors with GluN2C and GluN2D subunits Authors: Zhang M / Zhu S | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_33791.map.gz emd_33791.map.gz | 8.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-33791-v30.xml emd-33791-v30.xml emd-33791.xml emd-33791.xml | 21.9 KB 21.9 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_33791_fsc.xml emd_33791_fsc.xml | 10.3 KB | Display |  FSC data file FSC data file |
| Images |  emd_33791.png emd_33791.png | 119.5 KB | ||
| Filedesc metadata |  emd-33791.cif.gz emd-33791.cif.gz | 7.6 KB | ||
| Others |  emd_33791_half_map_1.map.gz emd_33791_half_map_1.map.gz emd_33791_half_map_2.map.gz emd_33791_half_map_2.map.gz | 72.8 MB 72.9 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-33791 http://ftp.pdbj.org/pub/emdb/structures/EMD-33791 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-33791 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-33791 | HTTPS FTP |
-Validation report
| Summary document |  emd_33791_validation.pdf.gz emd_33791_validation.pdf.gz | 723 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_33791_full_validation.pdf.gz emd_33791_full_validation.pdf.gz | 722.5 KB | Display | |
| Data in XML |  emd_33791_validation.xml.gz emd_33791_validation.xml.gz | 17.6 KB | Display | |
| Data in CIF |  emd_33791_validation.cif.gz emd_33791_validation.cif.gz | 23.3 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-33791 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-33791 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-33791 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-33791 | HTTPS FTP |
-Related structure data
| Related structure data |  7yfiMC  7yffC  7yfgC  7yfhC  7yflC  7yfmC  7yfoC  7yfrC  8hdkC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_33791.map.gz / Format: CCP4 / Size: 93 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_33791.map.gz / Format: CCP4 / Size: 93 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.071 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_33791_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_33791_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Rat tri-heteromeric GluN1-GluN2A-GluN2C NMDA receptor in complex ...
| Entire | Name: Rat tri-heteromeric GluN1-GluN2A-GluN2C NMDA receptor in complex with glycine and glutamate |
|---|---|
| Components |
|
-Supramolecule #1: Rat tri-heteromeric GluN1-GluN2A-GluN2C NMDA receptor in complex ...
| Supramolecule | Name: Rat tri-heteromeric GluN1-GluN2A-GluN2C NMDA receptor in complex with glycine and glutamate type: organelle_or_cellular_component / ID: 1 / Parent: 0 / Macromolecule list: #1-#3 |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Glutamate receptor ionotropic, NMDA 1
| Macromolecule | Name: Glutamate receptor ionotropic, NMDA 1 / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 89.956797 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MSTMHLLTFA LLFSCSFARA ACDPKIVNIG AVLSTRKHEQ MFREAVNQAN KRHGSWKIQL NATSVTHKPN AIQMALSVCE DLISSQVYA ILVSHPPTPN DHFTPTPVSY TAGFYRIPVL GLTTRMSIYS DKSIHLSFLR TVPPYSHQSS VWFEMMRVYN W NHIILLVS ...String: MSTMHLLTFA LLFSCSFARA ACDPKIVNIG AVLSTRKHEQ MFREAVNQAN KRHGSWKIQL NATSVTHKPN AIQMALSVCE DLISSQVYA ILVSHPPTPN DHFTPTPVSY TAGFYRIPVL GLTTRMSIYS DKSIHLSFLR TVPPYSHQSS VWFEMMRVYN W NHIILLVS DDHEGRAAQK RLETLLEERE SKAEKVLQFD PGTKNVTALL MEARELEARV IILSASEDDA ATVYRAAAML NM TGSGYVW LVGEREISGN ALRYAPDGII GLQLINGKNE SAHISDAVGV VAQAVHELLE KENITDPPRG CVGNTNIWKT GPL FKRVLM SSKYADGVTG RVEFNEDGDR KFANYSIMNL QNRKLVQVGI YNGTHVIPND RKIIWPGGET EKPRGYQMST RLKI VTIHQ EPFVYVKPTM SDGTCKEEFT VNGDPVKKVI CTGPNDTSPG SPRHTVPQCC YGFCIDLLIK LARTMNFTYE VHLVA DGKF GTQERVNNSN KKEWNGMMGE LLSGQADMIV APLTINNERA QYIEFSKPFK YQGLTILVKK EIPRSTLDSF MQPFQS TLW LLVGLSVHVV AVMLYLLDRF SPFGRFKVNS EEEEEDALTL SSAMWFSWGV LLNSGIGEGA PRSFSARILG MVWAGFA MI IVASYTANLA AFLVLDRPEE RITGINDPRL RNPSDKFIYA TVKQSSVDIY FRRQVELSTM YRHMEKHNYE SAAEAIQA V RDNKLHAFIW DSAVLEFEAS QKCDLVTTGE LFFRSGFGIG MRKDSPWKQN VSLSILKSHE NGFMEDLDKT WVRYQEC UniProtKB: Glutamate receptor ionotropic, NMDA 1 |
-Macromolecule #2: Glutamate receptor
| Macromolecule | Name: Glutamate receptor / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 93.837469 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: PMGRLGYWTL LVLPALLVWR DPAQNAAAEK GPPALNIAVL LGHSHDVTER ELRNLWGPEQ ATGLPLDVNV VALLMNRTDP KSLITHVCD LMSGARIHGL VFGDDTDQEA VAQMLDFISS QTFIPILGIH GGASMIMADK DPTSTFFQFG ASIQQQATVM L KIMQDYDW ...String: PMGRLGYWTL LVLPALLVWR DPAQNAAAEK GPPALNIAVL LGHSHDVTER ELRNLWGPEQ ATGLPLDVNV VALLMNRTDP KSLITHVCD LMSGARIHGL VFGDDTDQEA VAQMLDFISS QTFIPILGIH GGASMIMADK DPTSTFFQFG ASIQQQATVM L KIMQDYDW HVFSLVTTIF PGYRDFISFI KTTVDNSFVG WDMQNVITLD TSFEDAKTQV QLKKIHSSVI LLYCSKDEAV LI LSEARSL GLTGYDFFWI VPSLVSGNTE LIPKEFPSGL ISVSYDDWDY SLEARVRDGL GILTTAASSM LEKFSYIPEA KAS CYGQAE KPETPLHTLH QFMVNVTWDG KDLSFTEEGY QVHPRLVVIV LNKDREWEKV GKWENQTLSL RHAVWPRYKS FSDC EPDDN HLSIVTLEEA PFVIVEDIDP LTETCVRNTV PCRKFVKINN STNEGMNVKK CCKGFCIDIL KKLSRTVKFT YDLYL VTNG KHGKKVNNVW NGMIGEVVYQ RAVMAVGSLT INEERSEVVD FSVPFVETGI SVMVSRSNGT VSPSAFLEPF SASVWV MMF VMLLIVSAIA VFVFEYFSPV GYNRNLAKGK APHGPSFTIG KAIWLLWGLV FNNSVPVQNP KGTTSKIMVS VWAFFAV IF LASYTANLAA FMIQEEFVDQ VTGLSDKKFQ RPHDYSPPFR FGTVPNGSTE RNIRNNYPYM HQYMTRFNQR GVEDALVS L KTGKLDAFIY DAAVLNYKAG RDEGCKLVTI GSGYIFATTG YGIALQKGSP WKRQIDLALL QFVGDGEMEE LETLWLTGI CHNEKNEVMS SQLDIDNMAG VFYMLAAAMA LSLITFIW UniProtKB: Glutamate receptor |
-Macromolecule #3: Glutamate receptor ionotropic, NMDA 2C
| Macromolecule | Name: Glutamate receptor ionotropic, NMDA 2C / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 91.658445 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MGGALGPALL LTSLLGAWAR LGAGQGEQAV TVAVVFGSSG PLQTQARTRL TSQNFLDLPL EIQPLTVGVN NTNPSSILTQ ICGLLGAAR VHGIVFEDNV DTEAVAQLLD FVSSQTHVPI LSISGGSAVV LTPKEPGSAF LQLGVSLEQQ LQVLFKVLEE Y DWSAFAVI ...String: MGGALGPALL LTSLLGAWAR LGAGQGEQAV TVAVVFGSSG PLQTQARTRL TSQNFLDLPL EIQPLTVGVN NTNPSSILTQ ICGLLGAAR VHGIVFEDNV DTEAVAQLLD FVSSQTHVPI LSISGGSAVV LTPKEPGSAF LQLGVSLEQQ LQVLFKVLEE Y DWSAFAVI TSLHPGHALF LEGVRAVADA SYLSWRLLDV LTLELGPGGP RARTQRLLRQ VDAPVLVAYC SREEAEVLFA EA AQAGLVG PGHVWLVPNL ALGSTDAPPA AFPVGLISVV TESWRLSLRQ KVRDGVAILA LGAHSYRRQY GTLPAPAGDC RSH PGPVSP AREAFYRHLL NVTWEGRDFS FSPGGYLVRP TMVVIALNRH RLWEMVGRWD HGVLYMKYPV WPRYSTSLQP VVDS RHLTV ATLEERPFVI VESPDPGTGG CVPNTVPCRR QSNHTFSSGD LTPYTKLCCK GFCIDILKKL AKVVKFSYDL YLVTN GKHG KRVRGVWNGM IGEVYYKRAD MAIGSLTINE ERSEIIDFSV PFVETGISVM VSRSNGTVSP SAFLEPYSPA VWVMMF VMC LTVVAITVFM FEYFSPVSYN QNLTKGKKPG GPSFTIGKSV WLLWALVFNN SVPIENPRGT TSKIMVLVWA FFAVIFL AS YTANLAAFMI QEQYIDTVSG LSDKKFQRPQ DQYPPFRFGT VPNGSTERNI RSNYRDMHTH MVKFNQRSVE DALTSLKM G KLDAFIYDAA VLNYMAGKDE GCKLVTIGSG KVFATTGYGI AMQKDSHWKR AIDLALLQLL GDGETQKLET VWLSGICQN EKNEVMSSKL DIDNMAGVFY MLLVAMGLAL LVFAW UniProtKB: Glutamate receptor ionotropic, NMDA 2C |
-Macromolecule #6: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 6 / Number of copies: 21 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Macromolecule #7: GLYCINE
| Macromolecule | Name: GLYCINE / type: ligand / ID: 7 / Number of copies: 2 / Formula: GLY |
|---|---|
| Molecular weight | Theoretical: 75.067 Da |
| Chemical component information |  ChemComp-GLY: |
-Macromolecule #8: GLUTAMIC ACID
| Macromolecule | Name: GLUTAMIC ACID / type: ligand / ID: 8 / Number of copies: 2 / Formula: GLU |
|---|---|
| Molecular weight | Theoretical: 147.129 Da |
| Chemical component information |  ChemComp-GLU: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 4.2 mg/mL |
|---|---|
| Buffer | pH: 8 |
| Grid | Model: Quantifoil R1.2/1.3 / Material: GOLD / Mesh: 300 / Pretreatment - Type: GLOW DISCHARGE |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 281 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: DIRECT ELECTRON DE-10 (5k x 4k) / Digitization - Dimensions - Width: 5760 pixel / Digitization - Dimensions - Height: 9042 pixel / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.0 µm / Nominal defocus min: 1.2 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)