+Search query
-Structure paper
| Title | A Tail-Based Mechanism Drives Nucleosome Demethylation by the LSD2/NPAC Multimeric Complex. |
|---|---|
| Journal, issue, pages | Cell Rep, Vol. 27, Issue 2, Page 387-399.e7, Year 2019 |
| Publish date | Apr 9, 2019 |
 Authors Authors | Chiara Marabelli / Biagina Marrocco / Simona Pilotto / Sagar Chittori / Sarah Picaud / Sara Marchese / Giuseppe Ciossani / Federico Forneris / Panagis Filippakopoulos / Guy Schoehn / Daniela Rhodes / Sriram Subramaniam / Andrea Mattevi /       |
| PubMed Abstract | LSD1 and LSD2 are homologous histone demethylases with opposite biological outcomes related to chromatin silencing and transcription elongation, respectively. Unlike LSD1, LSD2 nucleosome-demethylase ...LSD1 and LSD2 are homologous histone demethylases with opposite biological outcomes related to chromatin silencing and transcription elongation, respectively. Unlike LSD1, LSD2 nucleosome-demethylase activity relies on a specific linker peptide from the multidomain protein NPAC. We used single-particle cryoelectron microscopy (cryo-EM), in combination with kinetic and mutational analysis, to analyze the mechanisms underlying the function of the human LSD2/NPAC-linker/nucleosome complex. Weak interactions between LSD2 and DNA enable multiple binding modes for the association of the demethylase to the nucleosome. The demethylase thereby captures mono- and dimethyl Lys4 of the H3 tail to afford histone demethylation. Our studies also establish that the dehydrogenase domain of NPAC serves as a catalytically inert oligomerization module. While LSD1/CoREST forms a nucleosome docking platform at silenced gene promoters, LSD2/NPAC is a multifunctional enzyme complex with flexible linkers, tailored for rapid chromatin modification, in conjunction with the advance of the RNA polymerase on actively transcribed genes. |
 External links External links |  Cell Rep / Cell Rep /  PubMed:30970244 PubMed:30970244 |
| Methods | SAS (X-ray synchrotron) / EM (single particle) |
| Resolution | 4.02 - 7.95 Å |
| Structure data | 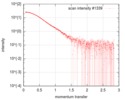 SASDFU3: Lysine-specific Demethylase (LSD2) (Lysyne-specific Demethylase LSD2, LSD2) 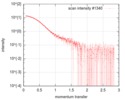 SASDFV3: Cytokine-like nuclear factor dehydrogenase domain, NPAC DH (NPAC delta-261) 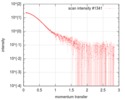 SASDFW3: Cytokine-like nuclear factor dehydrogenase domain, NPAC DH, plus native linker (NPAC delta-205) EMDB-4704, PDB-6r1t: EMDB-4705, PDB-6r1u: EMDB-4710, PDB-6r25:  EMDB-4711:  EMDB-4712: |
| Chemicals |  ChemComp-FAD:  ChemComp-ZN:  ChemComp-HOH: |
| Source |
|
 Keywords Keywords | GENE REGULATION / Histone demethylation / chromatin reader / flavoenzyme / epigenetics / evolution of protein function / molecular recognition. |
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About Yorodumi Papers
About Yorodumi Papers









 homo sapiens (human)
homo sapiens (human)