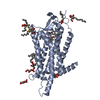
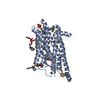
Title Comparison of Orexin 1 and Orexin 2 Ligand Binding Modes Using X-ray Crystallography and Computational Analysis. Journal, issue, pages J. Med. Chem. , Vol. 63, Page 1528-1543, Year 2020Publish date Dec 11, 2019 (structure data deposition date) Rappas, M. / Ali, A.A.E. / Bennett, K.A. / Brown, J.D. / Bucknell, S.J. / Congreve, M. / Cooke, R.M. / Cseke, G. / de Graaf, C. / Dore, A.S. ...Rappas, M. / Ali, A.A.E. / Bennett, K.A. / Brown, J.D. / Bucknell, S.J. / Congreve, M. / Cooke, R.M. / Cseke, G. / de Graaf, C. / Dore, A.S. / Errey, J.C. / Jazayeri, A. / Marshall, F.H. / Mason, J.S. / Mould, R. / Patel, J.C. / Tehan, B.G. / Weir, M. / Christopher, J.A. / Methods X-ray diffraction Resolution 2.11 - 3.04 Å Structure data PDB-6to7 suvorexant at 2.29 A resolutionMethod : X-RAY DIFFRACTION / Resolution : 2.26 Å
PDB-6tod EMPA Method : X-RAY DIFFRACTION / Resolution : 2.11 Å
PDB-6tos GSK1059865 Method : X-RAY DIFFRACTION / Resolution : 2.13 Å
PDB-6tot lemborexant Method : X-RAY DIFFRACTION / Resolution : 2.22 Å
PDB-6tp3 daridorexant Method : X-RAY DIFFRACTION / Resolution : 3.04 Å
PDB-6tp4 ACT-462206 Method : X-RAY DIFFRACTION / Resolution : 3.011 Å
PDB-6tp6 filorexant Method : X-RAY DIFFRACTION / Resolution : 2.338 Å
PDB-6tpg EMPA at 2.74 A resolutionMethod : X-RAY DIFFRACTION / Resolution : 2.741 Å
PDB-6tpj suvorexant at 2.76 A resolutionMethod : X-RAY DIFFRACTION / Resolution : 2.74 Å
PDB-6tpn HTL6641 at 2.61 A resolutionMethod : X-RAY DIFFRACTION / Resolution : 2.608 Å
PDB-6tq4 16 Method : X-RAY DIFFRACTION / Resolution : 2.299 Å
PDB-6tq6 14 Method : X-RAY DIFFRACTION / Resolution : 2.546 Å
PDB-6tq7 SB-334867 Method : X-RAY DIFFRACTION / Resolution : 2.6636 Å
PDB-6tq9 SB-408124 Method : X-RAY DIFFRACTION / Resolution : 2.655 Å
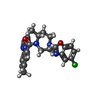
Chemicals ChemComp-SUV / medication, antagonist*YM
ChemComp-PG4 / precipitant*YM
ChemComp-SOG / detergent*YM
ChemComp-PGW / phospholipid*YM
ChemComp-7MA / antagonist*YM
ChemComp-NRE
ChemComp-NRK / medication, antagonist*YM
ChemComp-NS2 / medication, antagonist*YM
ChemComp-NRZ
ChemComp-NT5 / antagonist*YM
ChemComp-NU8 / HTL-6641*YM
ChemComp-NV8
ChemComp-NVH
ChemComp-NVK / antagonist, medication*YM
ChemComp-NVN / antagonist, medication*YM
Source homo sapiens (human)pyrococcus abyssi ge5 (archaea)pyrococcus abyssi (strain ge5 / orsay) (archaea) / /
 Authors
Authors External links
External links J. Med. Chem. /
J. Med. Chem. /  PubMed:31860301
PubMed:31860301





















 Keywords
Keywords Movie
Movie Controller
Controller Structure viewers
Structure viewers About Yorodumi Papers
About Yorodumi Papers



 homo sapiens (human)
homo sapiens (human)
 pyrococcus abyssi ge5 (archaea)
pyrococcus abyssi ge5 (archaea)