[English] 日本語
 Yorodumi
Yorodumi- PDB-6tos: Crystal structure of the Orexin-1 receptor in complex with GSK1059865 -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 6tos | ||||||
|---|---|---|---|---|---|---|---|
| Title | Crystal structure of the Orexin-1 receptor in complex with GSK1059865 | ||||||
 Components Components | Orexin receptor type 1 | ||||||
 Keywords Keywords | MEMBRANE PROTEIN / 7TM / GPCR | ||||||
| Function / homology |  Function and homology information Function and homology informationorexin receptor activity / Orexin and neuropeptides FF and QRFP bind to their respective receptors / feeding behavior / peptide hormone binding / regulation of cytosolic calcium ion concentration / neuropeptide signaling pathway / cellular response to hormone stimulus / G protein-coupled receptor activity / G alpha (q) signalling events / chemical synaptic transmission ...orexin receptor activity / Orexin and neuropeptides FF and QRFP bind to their respective receptors / feeding behavior / peptide hormone binding / regulation of cytosolic calcium ion concentration / neuropeptide signaling pathway / cellular response to hormone stimulus / G protein-coupled receptor activity / G alpha (q) signalling events / chemical synaptic transmission / positive regulation of ERK1 and ERK2 cascade / synapse / plasma membrane Similarity search - Function | ||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 2.13 Å MOLECULAR REPLACEMENT / Resolution: 2.13 Å | ||||||
 Authors Authors | Rappas, M. / Ali, A. / Bennett, K.A. / Brown, J.D. / Bucknell, S.J. / Congreve, M. / Cooke, R.M. / Cseke, G. / de Graaf, C. / Dore, A.S. ...Rappas, M. / Ali, A. / Bennett, K.A. / Brown, J.D. / Bucknell, S.J. / Congreve, M. / Cooke, R.M. / Cseke, G. / de Graaf, C. / Dore, A.S. / Errey, J.C. / Jazayeri, A. / Marshall, F.H. / Mason, J.S. / Mould, R. / Patel, J.C. / Tehan, B.G. / Weir, M. / Christopher, J.A. | ||||||
| Funding support |  United States, 1items United States, 1items
| ||||||
 Citation Citation |  Journal: J.Med.Chem. / Year: 2020 Journal: J.Med.Chem. / Year: 2020Title: Comparison of Orexin 1 and Orexin 2 Ligand Binding Modes Using X-ray Crystallography and Computational Analysis. Authors: Rappas, M. / Ali, A.A.E. / Bennett, K.A. / Brown, J.D. / Bucknell, S.J. / Congreve, M. / Cooke, R.M. / Cseke, G. / de Graaf, C. / Dore, A.S. / Errey, J.C. / Jazayeri, A. / Marshall, F.H. / ...Authors: Rappas, M. / Ali, A.A.E. / Bennett, K.A. / Brown, J.D. / Bucknell, S.J. / Congreve, M. / Cooke, R.M. / Cseke, G. / de Graaf, C. / Dore, A.S. / Errey, J.C. / Jazayeri, A. / Marshall, F.H. / Mason, J.S. / Mould, R. / Patel, J.C. / Tehan, B.G. / Weir, M. / Christopher, J.A. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  6tos.cif.gz 6tos.cif.gz | 285.1 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb6tos.ent.gz pdb6tos.ent.gz | 231.3 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  6tos.json.gz 6tos.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/to/6tos https://data.pdbj.org/pub/pdb/validation_reports/to/6tos ftp://data.pdbj.org/pub/pdb/validation_reports/to/6tos ftp://data.pdbj.org/pub/pdb/validation_reports/to/6tos | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  6to7SC  6todC  6totC  6tp3C  6tp4C  6tp6C  6tpgC  6tpjC  6tpnC  6tq4C  6tq6C  6tq7C  6tq9C S: Starting model for refinement C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 | 
| ||||||||
| 2 | 
| ||||||||
| Unit cell |
|
- Components
Components
-Protein / Sugars , 2 types, 29 molecules AB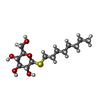

| #1: Protein | Mass: 38132.852 Da / Num. of mol.: 2 Mutation: E46A I85L V95A R162L N194A L198A Y211A L304V C339A C375W C376W Source method: isolated from a genetically manipulated source Details: GSK1059865 bound in the orthosteric site / Source: (gene. exp.)  Homo sapiens (human) / Gene: HCRTR1 / Production host: Homo sapiens (human) / Gene: HCRTR1 / Production host:  #6: Sugar | ChemComp-SOG / |
|---|
-Non-polymers , 7 types, 163 molecules 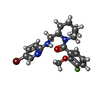











| #2: Chemical | | #3: Chemical | ChemComp-PG4 / | #4: Chemical | #5: Chemical | ChemComp-SO4 / #7: Chemical | ChemComp-NA / | #8: Chemical | #9: Water | ChemComp-HOH / | |
|---|
-Details
| Has ligand of interest | Y |
|---|---|
| Has protein modification | Y |
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 3.78 Å3/Da / Density % sol: 67.47 % |
|---|---|
| Crystal grow | Temperature: 284 K / Method: vapor diffusion, sitting drop / pH: 5 Details: 0.1M TRISODIUM CITRATE 50mM SODIUM CHLORIDE 50mM LITHIUM SULPHATE 15-34% PEG400 PH range: 3.0-6.5 / Temp details: Stable |
-Data collection
| Diffraction | Mean temperature: 100 K / Ambient temp details: Stable / Serial crystal experiment: N |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  Diamond Diamond  / Beamline: I24 / Wavelength: 0.98658 Å / Beamline: I24 / Wavelength: 0.98658 Å |
| Detector | Type: DECTRIS PILATUS3 6M / Detector: PIXEL / Date: Nov 25, 2015 |
| Radiation | Monochromator: Si (111) / Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 0.98658 Å / Relative weight: 1 |
| Reflection | Resolution: 2.13→30.47 Å / Num. obs: 44881 / % possible obs: 71.1 % / Redundancy: 3.3 % / CC1/2: 0.99 / Net I/σ(I): 8.8 |
| Reflection shell | Resolution: 2.13→2.25 Å / Num. unique obs: 898 / CC1/2: 0.338 / % possible all: 9.19 |
- Processing
Processing
| Software |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: 6TO7 Resolution: 2.13→30.47 Å / Cor.coef. Fo:Fc: 0.927 / Cor.coef. Fo:Fc free: 0.92 / SU R Cruickshank DPI: 0.232 / Cross valid method: THROUGHOUT / SU R Blow DPI: 0.222 / SU Rfree Blow DPI: 0.179 / SU Rfree Cruickshank DPI: 0.184
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 60.1 Å2
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine analyze | Luzzati coordinate error obs: 0.28 Å | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 2.13→30.47 Å
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell | Resolution: 2.13→2.25 Å
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement TLS params. | Refine-ID: X-RAY DIFFRACTION
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement TLS group |
|
 Movie
Movie Controller
Controller












 PDBj
PDBj














