-Search query
-Search result
Showing 1 - 50 of 679 items for (author: zeng & j)

EMDB-37249: 
Cryo-EM structure of EBV gH/gL-gp42 in complex with fab 2C1

PDB-8khr: 
Cryo-EM structure of EBV gH/gL-gp42 in complex with fab 2C1

EMDB-38203: 
Cryo-EM structure of HerA

EMDB-38204: 
Cryo-EM structure of an anti-phage defense complex

EMDB-38205: 
Cryo-EM structure of an anti-phage defense complex bound to AMPPNP and DNA at state 1

EMDB-38206: 
Cryo-EM structure of an anti-phage defense complex bound to AMPPNP and DNA at state 2

EMDB-38207: 
Cryo-EM structure of an anti-phage defense complex bound to ATPrS and DNA

PDB-8xau: 
Cryo-EM structure of HerA

PDB-8xav: 
Cryo-EM structure of an anti-phage defense complex

PDB-8xaw: 
Cryo-EM structure of an anti-phage defense complex bound to AMPPNP and DNA at state 1

PDB-8xax: 
Cryo-EM structure of an anti-phage defense complex bound to AMPPNP and DNA at state 2

PDB-8xay: 
Cryo-EM structure of an anti-phage defense complex bound to ATPrS and DNA

EMDB-41844: 
Cryo-EM structure of C.crescentus bNY30a pilus complex

EMDB-42136: 
PhiCb5 maturation protein with Caulobacter crescentus bNY30a pili

EMDB-42163: 
ssRNA phage PhiCb5 virion

PDB-8u2b: 
Cryo-EM structure of C.crescentus bNY30a pilus complex

PDB-8ucr: 
PhiCb5 maturation protein with Caulobacter crescentus bNY30a pili

PDB-8uej: 
ssRNA phage PhiCb5 virion

EMDB-36732: 
Cryo-EM structure of the gasdermin pore from Trichoplax adhaerens

EMDB-36733: 
Cryo-EM structure of the gasdermin pore from Trichoplax adhaerens

EMDB-36734: 
Cryo-EM structure of RCD-1 pore from Neurospora crassa

PDB-8jyw: 
Cryo-EM structure of the gasdermin pore from Trichoplax adhaerens

PDB-8jyz: 
Cryo-EM structure of RCD-1 pore from Neurospora crassa

EMDB-35939: 
The global structure of pre50S related to DbpA in state3

EMDB-40456: 
Cryo-EM structure of human NCX1 in complex with SEA0400
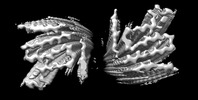
EMDB-36202: 
Cryo-EM structure of alpha-synuclein gS87 fibril

EMDB-36203: 
Cryo-EM structure of alpha-synuclein pS87 fibril

PDB-8jex: 
Cryo-EM structure of alpha-synuclein gS87 fibril

PDB-8jey: 
Cryo-EM structure of alpha-synuclein pS87 fibril

EMDB-35903: 
The global structure of pre50S related to DbpA in state1

EMDB-35905: 
The global structure of pre50S related to DbpA in state2

EMDB-35908: 
The global structure of pre50S related to DbpA in state4

EMDB-35910: 
The global structure of pre50S related to DbpA in state2

EMDB-35602: 
Cryo-EM structure of human HCN3 channel in apo state

PDB-8inz: 
Cryo-EM structure of human HCN3 channel in apo state

EMDB-41625: 
Type IV pilus from Pseudomonas PAO1 strain

EMDB-41632: 
Asymmetric reconstruction of mature PP7 virions

EMDB-41633: 
Type IV pilus from Pseudomonas PAO1 strain with PP7 Maturation protein
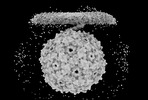
EMDB-41675: 
The original consensus map of PP7 binding to T4P from PAO1

EMDB-41780: 
Composite map of PP7 binding to type IV pilus from PAO1

PDB-8tum: 
Type IV pilus from Pseudomonas PAO1 strain

PDB-8tuw: 
Type IV pilus from Pseudomonas PAO1 strain with PP7 Maturation protein

PDB-8tux: 
Capsid of mature PP7 virion with 3'end region of PP7 genomic RNA

EMDB-41442: 
Acinetobacter GP16 Type IV pilus

EMDB-41443: 
Acinetobacter phage AP205

EMDB-41447: 
AP205 binding to one Acinetobacter GP16 type iv pilus

EMDB-41634: 
Inner Mat-T4P complex

EMDB-41635: 
Outer Mat-T4P complex

EMDB-41646: 
AP205 phage Acinetobacter gp16 T4P complex

EMDB-41657: 
Acinetobacter phage AP205 T=4 VLP
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
