-Search query
-Search result
Showing all 27 items for (author: zee & ct)

EMDB-0405: 
Amyloid-Beta (20-34) with L-isoaspartate 23
Method: electron crystallography / : Sawaya MR, Warmack RA

EMDB-20082: 
Amyloid-Beta (20-34) wild type
Method: electron crystallography / : Sawaya MR, Warmack RA

EMDB-7490: 
1.71 A MicroED structure of proteinase K at 0.86 e- / A^2
Method: electron crystallography / : Hattne J, Shi D

EMDB-7491: 
2.00 A MicroED structure of proteinase K at 2.6 e- / A^2
Method: electron crystallography / : Hattne J, Shi D

EMDB-7492: 
2.20 A MicroED structure of proteinase K at 4.3 e- / A^2
Method: electron crystallography / : Hattne J, Shi D

EMDB-7493: 
2.80 A MicroED structure of proteinase K at 6.0 e- / A^2
Method: electron crystallography / : Hattne J, Shi D

EMDB-7494: 
3.20 A MicroED structure of proteinase K at 7.8 e- / A^2
Method: electron crystallography / : Hattne J, Shi D

EMDB-7495: 
1.01 A MicroED structure of GSNQNNF at 0.27 e- / A^2
Method: electron crystallography / : Hattne J, Shi D

EMDB-7496: 
1.01 A MicroED structure of GSNQNNF at 0.81 e- / A^2
Method: electron crystallography / : Hattne J, Shi D
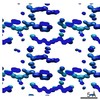
EMDB-7497: 
1.01 A MicroED structure of GSNQNNF at 1.3 e- / A^2
Method: electron crystallography / : Hattne J, Shi D

EMDB-7498: 
1.15 A MicroED structure of GSNQNNF at 1.9 e- / A^2
Method: electron crystallography / : Hattne J, Shi D

EMDB-7499: 
1.35 A MicroED structure of GSNQNNF at 2.4 e- / A^2
Method: electron crystallography / : Hattne J, Shi D

EMDB-7500: 
1.37 A MicroED structure of GSNQNNF at 2.9 e- / A^2
Method: electron crystallography / : Hattne J, Shi D

EMDB-7501: 
1.01 A MicroED structure of GSNQNNF at 0.17 e- / A^2
Method: electron crystallography / : Hattne J, Shi D

EMDB-7502: 
1.01 A MicroED structure of GSNQNNF at 0.50 e- / A^2
Method: electron crystallography / : Hattne J, Shi D

EMDB-7503: 
1.01 A MicroED structure of GSNQNNF at 0.82 e- / A^2
Method: electron crystallography / : Hattne J, Shi D

EMDB-7504: 
1.02 A MicroED structure of GSNQNNF at 1.2 e- / A^2
Method: electron crystallography / : Hattne J, Shi D

EMDB-7505: 
1.01 A MicroED structure of GSNQNNF at 1.5 e- / A^2
Method: electron crystallography / : Hattne J, Shi D
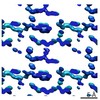
EMDB-7506: 
1.15 A MicroED structure of GSNQNNF at 1.8 e- / A^2
Method: electron crystallography / : Hattne J, Shi D
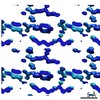
EMDB-7507: 
1.15 A MicroED structure of GSNQNNF at 2.1 e- / A^2
Method: electron crystallography / : Hattne J, Shi D

EMDB-7508: 
1.16 A MicroED structure of GSNQNNF at 2.5 e- / A^2
Method: electron crystallography / : Hattne J, Shi D
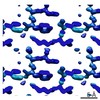
EMDB-7509: 
1.21 A MicroED structure of GSNQNNF at 2.8 e- / A^2
Method: electron crystallography / : Hattne J, Shi D
EMDB-7510: 
1.31 A MicroED structure of GSNQNNF at 3.1 e- / A^2
Method: electron crystallography / : Hattne J, Shi D
EMDB-7511: 
1.46 A MicroED structure of GSNQNNF at 3.4 e- / A^2
Method: electron crystallography / : Hattne J, Shi D
EMDB-7512: 
1.45 A MicroED structure of GSNQNNF at 3.8 e- / A^2
Method: electron crystallography / : Hattne J, Shi D

EMDB-7017: 
Segment from bank vole prion protein 168-176 QYNNQNNFV
Method: electron crystallography / : Glynn C, Rodriguez JA

EMDB-7287: 
MicroED structure of carbamazepine at 0.85 A resolution
Method: electron crystallography / : Gallagher-Jones M, Glynn C, Boyer DR, Martynowycz MW, Hernandez E, Miao J, Zee CT, NoviKova IV, Goldschmidt L, McFarlane HT, Helguera GF, Evans JE, Sawaya MR, Cascio D, Eisenberg D, Gonen T, Rodriguez JA
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
