+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-7287 | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | MicroED structure of carbamazepine at 0.85 A resolution | |||||||||||||||
 Map data Map data | 2Fo-Fc MicroED map of Carbamazepine (CBZ) | |||||||||||||||
 Sample Sample |
| |||||||||||||||
| Biological species | unidentified (others) | |||||||||||||||
| Method | electron crystallography / cryo EM / Resolution: 0.85 Å | |||||||||||||||
 Authors Authors | Gallagher-Jones M / Glynn C / Boyer DR / Martynowycz MW / Hernandez E / Miao J / Zee CT / NoviKova IV / Goldschmidt L / McFarlane HT ...Gallagher-Jones M / Glynn C / Boyer DR / Martynowycz MW / Hernandez E / Miao J / Zee CT / NoviKova IV / Goldschmidt L / McFarlane HT / Helguera GF / Evans JE / Sawaya MR / Cascio D / Eisenberg D / Gonen T / Rodriguez JA | |||||||||||||||
| Funding support |  United States, 4 items United States, 4 items
| |||||||||||||||
 Citation Citation |  Journal: Nat Struct Mol Biol / Year: 2018 Journal: Nat Struct Mol Biol / Year: 2018Title: Sub-ångström cryo-EM structure of a prion protofibril reveals a polar clasp. Authors: Marcus Gallagher-Jones / Calina Glynn / David R Boyer / Michael W Martynowycz / Evelyn Hernandez / Jennifer Miao / Chih-Te Zee / Irina V Novikova / Lukasz Goldschmidt / Heather T McFarlane / ...Authors: Marcus Gallagher-Jones / Calina Glynn / David R Boyer / Michael W Martynowycz / Evelyn Hernandez / Jennifer Miao / Chih-Te Zee / Irina V Novikova / Lukasz Goldschmidt / Heather T McFarlane / Gustavo F Helguera / James E Evans / Michael R Sawaya / Duilio Cascio / David S Eisenberg / Tamir Gonen / Jose A Rodriguez /   Abstract: The atomic structure of the infectious, protease-resistant, β-sheet-rich and fibrillar mammalian prion remains unknown. Through the cryo-EM method MicroED, we reveal the sub-ångström-resolution ...The atomic structure of the infectious, protease-resistant, β-sheet-rich and fibrillar mammalian prion remains unknown. Through the cryo-EM method MicroED, we reveal the sub-ångström-resolution structure of a protofibril formed by a wild-type segment from the β2-α2 loop of the bank vole prion protein. The structure of this protofibril reveals a stabilizing network of hydrogen bonds that link polar zippers within a sheet, producing motifs we have named 'polar clasps'. | |||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_7287.map.gz emd_7287.map.gz | 59 KB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-7287-v30.xml emd-7287-v30.xml emd-7287.xml emd-7287.xml | 18.7 KB 18.7 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_7287.png emd_7287.png | 262.2 KB | ||
| Others |  emd_7287_additional_1.map.gz emd_7287_additional_1.map.gz emd_7287_additional_2.map.gz emd_7287_additional_2.map.gz | 59 KB 58.8 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-7287 http://ftp.pdbj.org/pub/emdb/structures/EMD-7287 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-7287 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-7287 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_7287.map.gz / Format: CCP4 / Size: 68.4 KB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_7287.map.gz / Format: CCP4 / Size: 68.4 KB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | 2Fo-Fc MicroED map of Carbamazepine (CBZ) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. generated in cubic-lattice coordinate | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.281 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
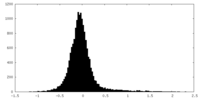
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 14 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Additional map: Fo MicroED map of Carbamazepine (CBZ)
| File | emd_7287_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Fo MicroED map of Carbamazepine (CBZ) | ||||||||||||
| Projections & Slices |
| ||||||||||||
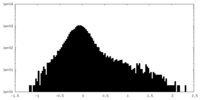
| Density Histograms |
-Additional map: Fo-Fc MicroED map of Carbamazepine (CBZ)
| File | emd_7287_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Fo-Fc MicroED map of Carbamazepine (CBZ) | ||||||||||||
| Projections & Slices |
| ||||||||||||
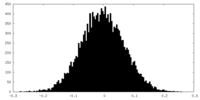
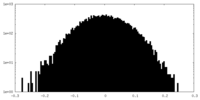
| Density Histograms |
- Sample components
Sample components
-Entire : Carbamazepine
| Entire | Name: Carbamazepine |
|---|---|
| Components |
|
-Supramolecule #1: Carbamazepine
| Supramolecule | Name: Carbamazepine / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 Details: Carbamazepine is a small molecule inhibitor of sodium channels used primarily in the treatment of epilepsy. |
|---|---|
| Source (natural) | Organism: unidentified (others) |
| Molecular weight | Theoretical: 234 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | electron crystallography |
| Aggregation state | 3D array |
- Sample preparation
Sample preparation
| Concentration | 1 mg/mL |
|---|---|
| Buffer | pH: 7.33 / Component - Concentration: 1.0 mg/mL / Component - Formula: C2H5OH / Component - Name: Ethanol / Details: Pure ethanol |
| Grid | Model: Quantifoil R2/4 / Material: COPPER / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY ARRAY / Support film - Film thickness: 10.0 nm / Pretreatment - Type: GLOW DISCHARGE |
| Vitrification | Cryogen name: NITROGEN / Chamber humidity: 30 % / Chamber temperature: 298 K / Instrument: FEI VITROBOT MARK IV / Details: Plunge-frozen after 10 seconds of blotting.. |
| Details | Powder was dissolved in ethanol at 1 mg/mL and applied to EM grids. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI F20 |
|---|---|
| Temperature | Min: 70.0 K / Max: 100.0 K |
| Image recording | Film or detector model: TVIPS TEMCAM-F416 (4k x 4k) / Digitization - Dimensions - Width: 4096 pixel / Digitization - Dimensions - Height: 4096 pixel / Number grids imaged: 1 / Number real images: 120 / Number diffraction images: 120 / Average exposure time: 5.0 sec. / Average electron dose: 0.01 e/Å2 Details: MicroED was collected under continuous rotation at a rate of 0.1 degrees/second with each frame corresponding to 5 seconds of exposure with a total range of tilts between -30 and +30 degrees. |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: DIFFRACTION / Camera length: 730 mm |
| Sample stage | Specimen holder model: GATAN 626 SINGLE TILT LIQUID NITROGEN CRYO TRANSFER HOLDER Cooling holder cryogen: NITROGEN / Tilt angle: -30.0, 30.0 |
| Experimental equipment |  Model: Tecnai F20 / Image courtesy: FEI Company |
- Image processing
Image processing
| Final reconstruction | Number classes used: 1 / Resolution.type: BY AUTHOR / Resolution: 0.85 Å / Resolution method: DIFFRACTION PATTERN/LAYERLINES |
|---|---|
| Crystal parameters | Unit cell - A: 7.5 Å / Unit cell - B: 11.0 Å / Unit cell - C: 13.70 Å / Unit cell - γ: 90 ° / Unit cell - α: 90 ° / Unit cell - β: 93.2 ° / Space group: P21/n |
| Crystallography statistics | Number intensities measured: 2598 / Number structure factors: 956 / Fourier space coverage: 49.4 / R sym: 0.105 / R merge: 0.086 / Overall phase error: 30 / Overall phase residual: 30 / Phase error rejection criteria: 0 / High resolution: 0.85 Å / Shell - Shell ID: 1 / Shell - High resolution: 0.85 Å / Shell - Low resolution: 7.5 Å / Shell - Number structure factors: 956 / Shell - Phase residual: 20 / Shell - Fourier space coverage: 49.4 / Shell - Multiplicity: 2.7 |
-Atomic model buiding 1
| Refinement | Space: RECIPROCAL / Protocol: AB INITIO MODEL / Overall B value: 10 / Target criteria: CC |
|---|
 Movie
Movie Controller
Controller







 Y (Sec.)
Y (Sec.) X (Row.)
X (Row.) Z (Col.)
Z (Col.)