-Search query
-Search result
Showing 1 - 50 of 89 items for (author: wu & sr)

EMDB-36071: 
Membrane bound PRTase, C3 symmetry, donor bound

EMDB-36072: 
Membrane bound PRTase, C3 symmetry, acceptor bound

PDB-8j8j: 
Membrane bound PRTase, C3 symmetry, donor bound

PDB-8j8k: 
Membrane bound PRTase, C3 symmetry, acceptor bound

EMDB-25427: 
Structure of KRAS G12V/HLA-A*03:01 in complex with antibody fragment V2

EMDB-35410: 
Arabinosyltransferase AftA

PDB-8if8: 
Arabinosyltransferase AftA

EMDB-35598: 
cryo-EM structure of the middle part of the shrimp white spot syndrome virus nucleocapsid (wide type)

EMDB-35600: 
Cryo-Em structure of the middle part of the shrimp white spot syndrome virus nucleocapsid (narrow type)
EMDB-29207: 
CryoET tomogram of mitochondria in BACHD mouse model neuron
EMDB-29208: 
CryoET tomogram of BACHD mouse model neuron showing sheet aggregates
EMDB-29210: 
CryoET tomogram of purified mitochondria from HD patient iPSC-derived neuron (Q109)
EMDB-29211: 
CryoET tomogram of HD patient iPSC-derived neuron (Q66) with PIAS1 hetKO treatment

EMDB-28668: 
CryoET tomogram of iPSC-derived control non-HD neuron (Q18)
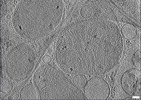
EMDB-28944: 
CryoET tomogram of iPSC-derived control non-HD neuron (Q20)
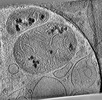
EMDB-28946: 
CryoET tomogram of HD patient iPSC-derived neuron (Q53)
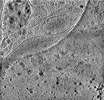
EMDB-29074: 
CryoET tomogram of HD patient iPSC-derived neuron (Q66)

EMDB-29075: 
CryoET tomogram of HD patient iPSC-derived neuron (Q77)

EMDB-29076: 
CryoET tomogram of HD patient iPSC-derived neuron (Q109)

EMDB-29079: 
CryoET tomogram of HD patient iPSC-derived neuron (Q66) showing sheet aggregate

EMDB-29080: 
CryoET tomogram of HD patient iPSC-derived neuron (Q66) with PIAS1 hetKO treatment
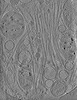
EMDB-29081: 
CryoET tomogram of HD patient iPSC-derived neuron (Q66) with GRSF1 KD treatment

EMDB-29083: 
CryoET tomogram of mitochondria in WT mouse neuron

EMDB-29084: 
CryoET tomogram of mitochondria in BACHD-dN17 mouse model neuron

EMDB-14860: 
Cryo-EM structure of the MVV CSC intasome at 4.5A resolution

EMDB-32832: 
SARS-CoV-2 Spike in complex with Fab of m31A7

PDB-7wuh: 
SARS-CoV-2 Spike in complex with Fab of m31A7

EMDB-32825: 
Negative stain volume of the mono-GlcNAc-decorated SARS-CoV-2 Spike

EMDB-23302: 
Cryo-EM structure of NLRP3 double-ring cage, 6-fold (12-mer)

EMDB-23303: 
Cryo-EM structure of NLRP3 double-ring cage, 6-fold (12-mer)

EMDB-23304: 
Cryo-EM structure of NLRP3 double-ring cage, 7-fold (14-mer)

EMDB-23305: 
Cryo-EM structure of NLRP3 double-ring cage, 8-fold (16-mer)

PDB-7lfh: 
Cryo-EM structure of NLRP3 double-ring cage, 6-fold (12-mer)

EMDB-30381: 
Cryo-EM density of SARS-CoV-2 spike protein

EMDB-31470: 
Cryo-EM structure of SARS-CoV-2 spike in complex with a neutralizing antibody chAb-25 (Focused refinement of S-RBD and chAb-25 region)

EMDB-31471: 
Cryo-EM structure of SARS-CoV-2 spike in complex with a neutralizing antibody chAb-45 (Focused refinement of S-RBD and chAb-45 region)

PDB-7f62: 
Cryo-EM structure of SARS-CoV-2 spike in complex with a neutralizing antibody chAb-25 (Focused refinement of S-RBD and chAb-25 region)

PDB-7f63: 
Cryo-EM structure of SARS-CoV-2 spike in complex with a neutralizing antibody chAb-45 (Focused refinement of S-RBD and chAb-45 region)

EMDB-23099: 
Structure of NTS-NTSR1-Gi complex in lipid nanodisc, canonical state, AHD and nanodisc mask out

EMDB-23100: 
Structure of NTS-NTSR1-Gi complex in lipid nanodisc, canonical state, overall

EMDB-23101: 
Structure of NTS-NTSR1-Gi complex in lipid nanodisc, noncanonical state, AHD and nanodisc mask out

EMDB-23102: 
Structure of NTS-NTSR1-Gi complex in lipid nanodisc, noncanonical state, overall

PDB-7l0p: 
Structure of NTS-NTSR1-Gi complex in lipid nanodisc, canonical state, without AHD

PDB-7l0q: 
Structure of NTS-NTSR1-Gi complex in lipid nanodisc, canonical state, with AHD

PDB-7l0r: 
Structure of NTS-NTSR1-Gi complex in lipid nanodisc, noncanonical state, without AHD

PDB-7l0s: 
Structure of NTS-NTSR1-Gi complex in lipid nanodisc, noncanonical state, with AHD

EMDB-30380: 
Negative stain density of the tetrameric FRIL in solution

EMDB-30419: 
Cryo-EM structure of SARS-CoV-2 Spike ectodomain

PDB-7cn9: 
Cryo-EM structure of SARS-CoV-2 Spike ectodomain

EMDB-30216: 
Cryo-EM structure of Mycobacterium smegmatis arabinosyltransferase EmbA-EmbB-AcpM2 in complex with ethambutol
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
