-Search query
-Search result
Showing 1 - 50 of 320 items for (author: todd & t)

EMDB-18878: 
Cryo-EM structure of the Asgard archaeal Argonaute HrAgo1 bound to a guide RNA

PDB-8r3z: 
Cryo-EM structure of the Asgard archaeal Argonaute HrAgo1 bound to a guide RNA

EMDB-41409: 
Cryo-EM structure of PCSK9 mimic HIT01-K21Q-R218E with AMG145 Fab
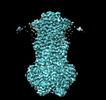
EMDB-40180: 
MsbA bound to cerastecin C

PDB-8gk7: 
MsbA bound to cerastecin C

EMDB-40260: 
CryoEM map of a de novo designed T=4 icosahedral nanocage hierarchically built from pseudosymmetric trimers; design Ico(T=4)-4

EMDB-40267: 
CryoEM map of a T=1 off-target state of design Ico(T=4)-4

EMDB-40268: 
CryoEM map of a de novo designed T=4 octahedral nanocage hierarchically built from pseudosymmetric trimers; design Oct(T=4)-3

EMDB-40269: 
CryoEM map of a T=1 off-target state of design Oct(T=4)-3

EMDB-40208: 
Backbone model of de novo-designed chlorophyll-binding nanocage O32-15

EMDB-40209: 
Chlorophyll-binding region of de novo-designed nanocage O32-15

PDB-8glt: 
Backbone model of de novo-designed chlorophyll-binding nanocage O32-15
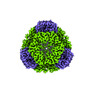
EMDB-42286: 
T33-ml28 - Designed Tetrahedral Protein Cage Using Machine Learning Algorithms
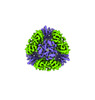
EMDB-42355: 
T33-ml30 - Designed Tetrahedral Protein Cage Using Machine Learning Algorithms
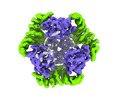
EMDB-42382: 
T33-ml35 Assembly Intermediate - Designed Tetrahedral Protein Cage Using Machine Learning Algorithms
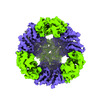
EMDB-42390: 
T33-ml23 Assembly Intermediate - Designed Tetrahedral Protein Cage Using Machine Learning Algorithms

PDB-8ui2: 
T33-ml28 - Designed Tetrahedral Protein Cage Using Machine Learning Algorithms

PDB-8ukm: 
T33-ml30 - Designed Tetrahedral Protein Cage Using Machine Learning Algorithms

PDB-8umr: 
T33-ml35 Assembly Intermediate - Designed Tetrahedral Protein Cage Using Machine Learning Algorithms

PDB-8un1: 
T33-ml23 Assembly Intermediate - Designed Tetrahedral Protein Cage Using Machine Learning Algorithms

PDB-8t1n: 
Micro-ED Structure of a Novel Domain of Unknown Function Solved with AlphaFold

EMDB-41048: 
Lassa GPC Trimer in complex with Fab 8.11G and nanobody D5

PDB-8t5c: 
Lassa GPC Trimer in complex with Fab 8.11G and nanobody D5

EMDB-42181: 
T33-ml23 - Designed Tetrahedral Protein Cage Using Machine Learning Algorithms
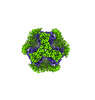
EMDB-42381: 
T33-ml35 - Designed Tetrahedral Protein Cage Using Machine Learning Algorithms

PDB-8uf0: 
T33-ml23 - Designed Tetrahedral Protein Cage Using Machine Learning Algorithms

PDB-8ump: 
T33-ml35 - Designed Tetrahedral Protein Cage Using Machine Learning Algorithms

EMDB-28743: 
Cryo-EM structure of the S. cerevisiae Arf-like protein Arl1 bound to the Arf guanine nucleotide exchange factor Gea2

EMDB-28744: 
Cryo-EM consensus map of the S. cerevisiae Arf-like protein Arl1 bound to its effector guanine nucleotide exchange factor Gea2

EMDB-28746: 
Cryo-EM local map covering Gea2 protomer B HDS domains of the S. cerevisiae Arf-like protein Arl1 bound to the Arf guanine nucleotide exchange factor Gea2

EMDB-28747: 
Cryo-EM local map covering Gea2 protomer A HDS domains of the S. cerevisiae Arf-like protein Arl1 bound to the Arf guanine nucleotide exchange factor Gea2

EMDB-28748: 
Cryo-EM structure of the S. cerevisiae guanine nucleotide exchange factor Gea2

EMDB-28749: 
Cryo-EM consensus map of the S. cerevisiae guanine nucleotide exchange factor Gea2

EMDB-28750: 
Cryo-EM local map covering Gea2 protomer B HDS domains of the S. cerevisiae guanine nucleotide exchange factor Gea2

EMDB-28751: 
Cryo-EM local map covering Gea2 protomer A HDS domains of the S. cerevisiae guanine nucleotide exchange factor Gea2

PDB-8ezj: 
Cryo-EM structure of the S. cerevisiae Arf-like protein Arl1 bound to the Arf guanine nucleotide exchange factor Gea2

PDB-8ezq: 
Cryo-EM structure of the S. cerevisiae guanine nucleotide exchange factor Gea2

EMDB-41302: 
Lassa GPC trimer in complex with Fab GP23

EMDB-29700: 
Cryo-EM imaging scaffold subunits A and B used to display KRAS G12C complex with GDP

EMDB-29713: 
KRAS G12C complex with GDP imaged on a cryo-EM imaging scaffold

EMDB-29715: 
KRAS G12C complex with GDP and AMG 510 imaged on a cryo-EM imaging scaffold

EMDB-29718: 
Green Fluorescence Protein imaged on a cryo-EM imaging scaffold

EMDB-29719: 
KRAS G12V complex with GDP imaged on a cryo-EM imaging scaffold
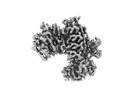
EMDB-29720: 
KRAS G13C complex with GDP imaged on a cryo-EM imaging scaffold

PDB-8g3k: 
Cryo-EM imaging scaffold subunits A and B used to display KRAS G12C complex with GDP

PDB-8g42: 
KRAS G12C complex with GDP imaged on a cryo-EM imaging scaffold

PDB-8g47: 
KRAS G12C complex with GDP and AMG 510 imaged on a cryo-EM imaging scaffold

PDB-8g4e: 
Green Fluorescence Protein imaged on a cryo-EM imaging scaffold

PDB-8g4f: 
KRAS G12V complex with GDP imaged on a cryo-EM imaging scaffold

PDB-8g4h: 
KRAS G13C complex with GDP imaged on a cryo-EM imaging scaffold
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
