[English] 日本語
 Yorodumi
Yorodumi- EMDB-28744: Cryo-EM consensus map of the S. cerevisiae Arf-like protein Arl1 ... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM consensus map of the S. cerevisiae Arf-like protein Arl1 bound to the Arf guanine nucleotide exchange factor Gea2 | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | small GTPase / guanine nucleotide exchange factor / membrane trafficking / lipid flippase / trans-Golgi network / protein transport / LIPID TRANSPORT | |||||||||
| Biological species |  | |||||||||
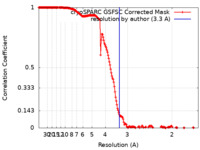
| Method | single particle reconstruction / cryo EM / Resolution: 3.3 Å | |||||||||
 Authors Authors | Duan HD / Li H | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2024 Journal: Nat Commun / Year: 2024Title: Structural insight into an Arl1-ArfGEF complex involved in Golgi recruitment of a GRIP-domain golgin. Authors: H Diessel Duan / Bhawik K Jain / Hua Li / Todd R Graham / Huilin Li /  Abstract: Arl1 is an Arf-like (Arl) GTP-binding protein that interacts with the guanine nucleotide exchange factor Gea2 to recruit the golgin Imh1 to the Golgi. The Arl1-Gea2 complex also binds and activates ...Arl1 is an Arf-like (Arl) GTP-binding protein that interacts with the guanine nucleotide exchange factor Gea2 to recruit the golgin Imh1 to the Golgi. The Arl1-Gea2 complex also binds and activates the phosphatidylserine flippase Drs2 and these functions may be related, although the underlying molecular mechanism is unclear. Here we report high-resolution cryo-EM structures of the full-length Gea2 and the Arl1-Gea2 complex. Gea2 is a large protein with 1459 residues and is composed of six domains (DCB, HUS, SEC7, HDS1-3). We show that Gea2 assembles a stable dimer via an extensive interface involving hydrophobic and electrostatic interactions in the DCB and HUS region. Contrary to the previous report on a Gea2 homolog in which Arl1 binds to the dimerization surface of the DCB domain, implying a disrupted dimer upon Arl1 binding, we find that Arl1 binds to the outside surface of the Gea2 DCB domain, leaving the Gea2 dimer intact. The interaction between Arl1 and Gea2 involves the classic FWY aromatic residue triad as well as two Arl1-specific residues. We show that key mutations that disrupt the Arl1-Gea2 interaction abrogate Imh1 Golgi association. This work clarifies the Arl1-Gea2 interaction and improves our understanding of molecular events in the membrane trafficking. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_28744.map.gz emd_28744.map.gz | 427.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-28744-v30.xml emd-28744-v30.xml emd-28744.xml emd-28744.xml | 15.2 KB 15.2 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_28744_fsc.xml emd_28744_fsc.xml | 17 KB | Display |  FSC data file FSC data file |
| Images |  emd_28744.png emd_28744.png | 49.3 KB | ||
| Filedesc metadata |  emd-28744.cif.gz emd-28744.cif.gz | 5.9 KB | ||
| Others |  emd_28744_half_map_1.map.gz emd_28744_half_map_1.map.gz emd_28744_half_map_2.map.gz emd_28744_half_map_2.map.gz | 474.6 MB 474.6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-28744 http://ftp.pdbj.org/pub/emdb/structures/EMD-28744 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-28744 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-28744 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_28744.map.gz / Format: CCP4 / Size: 512 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_28744.map.gz / Format: CCP4 / Size: 512 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.828 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_28744_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_28744_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Gea2 homodimer in complex with two Arl1 monomers
| Entire | Name: Gea2 homodimer in complex with two Arl1 monomers |
|---|---|
| Components |
|
-Supramolecule #1: Gea2 homodimer in complex with two Arl1 monomers
| Supramolecule | Name: Gea2 homodimer in complex with two Arl1 monomers / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: ARF guanine-nucleotide exchange factor 2
| Macromolecule | Name: ARF guanine-nucleotide exchange factor 2 / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Recombinant expression | Organism:  |
| Sequence | String: MHHHHHHSSG VDLGTENLYF QSNAMSDREF VTVDPVTIII KECINLSTAM RKYSKFTSQS GVAALLGGGS EIFSNQDDYL AHTFNNLNT NKHNDPFLSG FIQLRLMLNK LKNLDNIDSL TILQPFLLIV STSSISGYIT SLALDSLQKF FTLNIINESS Q NYIGAHRA ...String: MHHHHHHSSG VDLGTENLYF QSNAMSDREF VTVDPVTIII KECINLSTAM RKYSKFTSQS GVAALLGGGS EIFSNQDDYL AHTFNNLNT NKHNDPFLSG FIQLRLMLNK LKNLDNIDSL TILQPFLLIV STSSISGYIT SLALDSLQKF FTLNIINESS Q NYIGAHRA TVNALTHCRF EGSQQLSDDS VLLKVVFLLR SIVDSPYGDL LSNSIIYDVL QTILSLACNN RRSEVLRNAA QS TMIAVTV KIFSKLKTIE PVNVNQIYIN DESYTNDVLK ADTIGTNVES KEEGSQEDPI GMKVNNEEAI SEDDGIEEEH IHS EKSTNG AEQLDIVQKT TRSNSRIQAY ADDNYGLPVV RQYLNLLLSL IAPENELKHS YSTRIFGLEL IQTALEISGD RLQL YPRLF TLISDPIFKS ILFIIQNTTK LSLLQATLQL FTTLVVILGN NLQLQIELTL TRIFSILLDD GTANNSSSEN KNKPS IIKE LLIEQISILW TRSPSFFTST FINFDCNLDR ADVSINFLKA LTKLALPESA LTTTESVPPI CLEGLVSLVD DMFDHM KDI DREEFGRQKN EMEILKKRDR KTEFIECTNA FNEKPKKGIP MLIEKGFIAS DSDKDIAEFL FNNNNRMNKK TIGLLLC HP DKVSLLNEYI RLFDFSGLRV DEAIRILLTK FRLPGESQQI ERIIEAFSSA YCENQDYDPS KISDNAEDDI STVQPDAD S VFILSYSIIM LNTDLHNPQV KEHMSFEDYS GNLKGCCNHK DFPFWYLDRI YCSIRDKEIV MPEEHHGNEK WFEDAWNNL ISSTTVITEI KKDTQSVMDK LTPLELLNFD RAIFKQVGPS IVSTLFNIYV VASDDHISTR MITSLDKCSY ISAFFDFKDL FNDILNSIA KGTTLINSSH DDELSTLAFE YGPMPLVQIK FEDTNTEIPV STDAVRFGRS FKGQLNTVVF FRIIRRNKDP K IFSKELWL NIVNIILTLY EDLILSPDIF PDLQKRLKLS NLPKPSPEIS INKSKESKGL LSTFASYLKG DEEPTEEEIK SS KKAMECI KSSNIAASVF GNESNITADL IKTLLDSAKT EKNADNSRYF EAELLFIIEL TIALFLFCKE EKELGKFILQ KVF QLSHTK GLTKRTVRRM LTYKILLISL CADQTEYLSK LINDELLKKG DIFTQKFFAT NQGKEFLKRL FSLTESEFYR GFLL GNENF WKFLRKVTAM KEQSESIFEY LNESIKTDSN ILTNENFMWV LGLLDEISSM GAVGNHWEIE YKKLTESGHK IDKEN PYKK SIELSLKSIQ LTSHLLEDNN DLRKNEIFAI IQALAHQCIN PCKQISEFAV VTLEQTLINK IEIPTNEMES VEELIE GGL LPLLNSSETQ EDQKILISSI LTIISNVYLH YLKLGKTSNE TFLKILSIFN KFVEDSDIEK KLQQLILDKK SIEKGNG SS SHGSAHEQTP ESNDVEIEAT APIDDNTDDD NKPKLSDVEK D |
-Macromolecule #2: ADP-ribosylation factor-like protein 1
| Macromolecule | Name: ADP-ribosylation factor-like protein 1 / type: protein_or_peptide / ID: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Recombinant expression | Organism:  |
| Sequence | String: ELRILILGLD GAGKTTILYR LQIGEVVTTK PTIGFNVETL SYKNLKLNVW DLGGLTSIRP YWRCYYADTA AVIFVVDSTD KDRMSTASK ELHLMLQEEE LQDAALLVFA NKQDQPGALS ASEVSKELNL VELKDRSWSI VASSAIKGEG ITEGLDWLID V IKEEQLAG ENLYFQSAGH HHHHH |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 69.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 1.7 µm / Nominal defocus min: 1.3 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller












 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)