+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | CryoEM map of a T=1 off-target state of design Ico(T=4)-4 | |||||||||
 Map data Map data | sharpened map from deepEMhancer | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | icosahedral nanocage / T=4 / T=4 icosahedra / T=1 / T=1 icosahedra / off-target / nanomaterial / computational design / de novo / DE NOVO PROTEIN | |||||||||
| Biological species | synthetic construct (others) | |||||||||
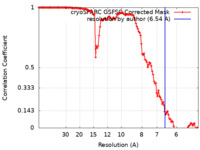
| Method | single particle reconstruction / cryo EM / Resolution: 6.54 Å | |||||||||
 Authors Authors | Borst AJ / Kibler RD / Lee S | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Nature / Year: 2025 Journal: Nature / Year: 2025Title: Four-component protein nanocages designed by programmed symmetry breaking. Authors: Sangmin Lee / Ryan D Kibler / Green Ahn / Yang Hsia / Andrew J Borst / Annika Philomin / Madison A Kennedy / Buwei Huang / Barry Stoddard / David Baker /   Abstract: Four, eight or twenty C3 symmetric protein trimers can be arranged with tetrahedral, octahedral or icosahedral point group symmetry to generate closed cage-like structures. Viruses access more ...Four, eight or twenty C3 symmetric protein trimers can be arranged with tetrahedral, octahedral or icosahedral point group symmetry to generate closed cage-like structures. Viruses access more complex higher triangulation number icosahedral architectures by breaking perfect point group symmetry, but nature appears not to have explored similar symmetry breaking for tetrahedral or octahedral symmetries. Here we describe a general design strategy for building higher triangulation number architectures starting from regular polyhedra through pseudosymmetrization of trimeric building blocks. Electron microscopy confirms the structures of T = 4 cages with 48 (tetrahedral), 96 (octahedral) and 240 (icosahedral) subunits, each with 4 distinct chains and 6 different protein-protein interfaces, and diameters of 33 nm, 43 nm and 75 nm, respectively. Higher triangulation number viruses possess very sophisticated functionalities; our general route to higher triangulation number nanocages should similarly enable a next generation of multiple antigen-displaying vaccine candidates and targeted delivery vehicles. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_40267.map.gz emd_40267.map.gz | 165 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-40267-v30.xml emd-40267-v30.xml emd-40267.xml emd-40267.xml | 15.2 KB 15.2 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_40267_fsc.xml emd_40267_fsc.xml | 11.9 KB | Display |  FSC data file FSC data file |
| Images |  emd_40267.png emd_40267.png | 154 KB | ||
| Filedesc metadata |  emd-40267.cif.gz emd-40267.cif.gz | 4 KB | ||
| Others |  emd_40267_additional_1.map.gz emd_40267_additional_1.map.gz emd_40267_half_map_1.map.gz emd_40267_half_map_1.map.gz emd_40267_half_map_2.map.gz emd_40267_half_map_2.map.gz | 89.5 MB 165 MB 165 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-40267 http://ftp.pdbj.org/pub/emdb/structures/EMD-40267 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-40267 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-40267 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_40267.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_40267.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | sharpened map from deepEMhancer | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2.557 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: unsharpened map from final Non-Uniform Refinement
| File | emd_40267_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | unsharpened map from final Non-Uniform Refinement | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: half map A from final Non-Uniform Refinement
| File | emd_40267_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map A from final Non-Uniform Refinement | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: half map B from final Non-Uniform Refinement
| File | emd_40267_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map B from final Non-Uniform Refinement | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Icos(T=4)-4
| Entire | Name: Icos(T=4)-4 |
|---|---|
| Components |
|
-Supramolecule #1: Icos(T=4)-4
| Supramolecule | Name: Icos(T=4)-4 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#4 / Details: A hierarchically designed T=4 icosahedral nanocage |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 / Details: 25mM Tris, 300mM NaCl |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Number real images: 6678 / Average electron dose: 61.463 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 1.7 µm / Nominal defocus min: 0.8 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller








 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)