-Search query
-Search result
Showing all 38 items for (author: rosenberg & os)

EMDB-41195: 
Tropomyosin-receptor kinase fused gene protein (TRK-fused gene protein; TFG) Low Complexity Domain (residues 237-327) G269V mutant, amyloid fiber
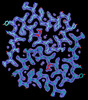
EMDB-41198: 
Tropomyosin-receptor kinase fused gene protein (TRK-fused gene protein; TFG) Low Complexity Domain (residues 237-327) P285L mutant, amyloid fiber

PDB-8teq: 
Tropomyosin-receptor kinase fused gene protein (TRK-fused gene protein; TFG) Low Complexity Domain (residues 237-327) G269V mutant, amyloid fiber

PDB-8ter: 
Tropomyosin-receptor kinase fused gene protein (TRK-fused gene protein; TFG) Low Complexity Domain (residues 237-327) P285L mutant, amyloid fiber
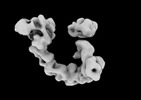
EMDB-29101: 
EGFR:Degrader:VHL:Elongin-B/C:Cul2

EMDB-26574: 
KS-AT di-domain of mycobacterial Pks13 with endogenous KS ligand bound

EMDB-27002: 
ACP1-KS-AT domains of mycobacterial Pks13

EMDB-27003: 
KS-AT domains of mycobacterial Pks13 with inward AT conformation

EMDB-27004: 
KS-AT domains of mycobacterial Pks13 with outward AT conformation

EMDB-27005: 
ACP1-KS-AT domains of mycobacterial Pks13

PDB-7uk4: 
KS-AT di-domain of mycobacterial Pks13 with endogenous KS ligand bound

PDB-8cuy: 
ACP1-KS-AT domains of mycobacterial Pks13

PDB-8cuz: 
KS-AT domains of mycobacterial Pks13 with inward AT conformation

PDB-8cv0: 
KS-AT domains of mycobacterial Pks13 with outward AT conformation

PDB-8cv1: 
ACP1-KS-AT domains of mycobacterial Pks13

EMDB-27730: 
SARS-CoV-2 Wuhan-hu-1-Spike-RBD bound to linker variant of affinity matured ACE2 mimetic CVD432

EMDB-27731: 
SARS-CoV-2 Wuhan-hu-1-Spike-RBD bound to computationally engineered ACE2 mimetic CVD293

PDB-8dv1: 
SARS-CoV-2 Wuhan-hu-1-Spike-RBD bound to linker variant of affinity matured ACE2 mimetic CVD432

PDB-8dv2: 
SARS-CoV-2 Wuhan-hu-1-Spike-RBD bound to computationally engineered ACE2 mimetic CVD293

EMDB-23970: 
Full length SARS-CoV-2 Nsp2

EMDB-23971: 
SARS-CoV-2 Nsp2

PDB-7msw: 
Full length SARS-CoV-2 Nsp2

PDB-7msx: 
SARS-CoV-2 Nsp2

EMDB-22829: 
Human Tom70 in complex with SARS CoV2 Orf9b

PDB-7kdt: 
Human Tom70 in complex with SARS CoV2 Orf9b

EMDB-20820: 
A complete structure of the ESX-3 translocon complex

PDB-6umm: 
A complete structure of the ESX-3 translocon complex

EMDB-20282: 
Inactive State of Manduca sexta soluble guanylate cyclase

EMDB-20283: 
Active State of Manduca sexta soluble Guanylate Cyclase

PDB-6pas: 
Inactive State of Manduca sexta soluble guanylate cyclase

PDB-6pat: 
Active State of Manduca sexta soluble Guanylate Cyclase

EMDB-20266: 
Cryo-EM structure of LbCas12a-crRNA: AcrVA4 (1:2 complex)

EMDB-20267: 
Cryo-EM structure of LbCas12a-crRNA: AcrVA4 (2:2 complex)

PDB-6p7m: 
Cryo-EM structure of LbCas12a-crRNA: AcrVA4 (1:2 complex)

PDB-6p7n: 
Cryo-EM structure of LbCas12a-crRNA: AcrVA4 (2:2 complex)

EMDB-2715: 
Electron cryo-microscopy of ABCG2 from two-dimensional crystals

PDB-4a82: 
Fitted model of staphylococcus aureus sav1866 model ABC transporter in the human cystic fibrosis transmembrane conductance regulator volume map EMD-1966.

EMDB-1966: 
CFTR map generated from 2D crystals grown using the epitaxial method.
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
