+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-20267 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of LbCas12a-crRNA: AcrVA4 (2:2 complex) | |||||||||
 Map data Map data | 3D Reconstruction (State II) | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | CRISPR-Cas / anti-CRISPR / Cas12a / Cpf1 / LbCas12a / AcrVA4 / RNA BINDING PROTEIN-RNA complex | |||||||||
| Function / homology |  Function and homology information Function and homology information | |||||||||
| Biological species |  Lachnospiraceae bacterium ND2006 (bacteria) / Lachnospiraceae bacterium ND2006 (bacteria) /  Moraxella bovoculi (bacteria) Moraxella bovoculi (bacteria) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 4.9 Å | |||||||||
 Authors Authors | Knott GJ / Liu JJ | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation | Journal: Acta Crystallogr D Biol Crystallogr / Year: 2010 Title: PHENIX: a comprehensive Python-based system for macromolecular structure solution. Authors: Paul D Adams / Pavel V Afonine / Gábor Bunkóczi / Vincent B Chen / Ian W Davis / Nathaniel Echols / Jeffrey J Headd / Li-Wei Hung / Gary J Kapral / Ralf W Grosse-Kunstleve / Airlie J McCoy ...Authors: Paul D Adams / Pavel V Afonine / Gábor Bunkóczi / Vincent B Chen / Ian W Davis / Nathaniel Echols / Jeffrey J Headd / Li-Wei Hung / Gary J Kapral / Ralf W Grosse-Kunstleve / Airlie J McCoy / Nigel W Moriarty / Robert Oeffner / Randy J Read / David C Richardson / Jane S Richardson / Thomas C Terwilliger / Peter H Zwart /  Abstract: Macromolecular X-ray crystallography is routinely applied to understand biological processes at a molecular level. However, significant time and effort are still required to solve and complete many ...Macromolecular X-ray crystallography is routinely applied to understand biological processes at a molecular level. However, significant time and effort are still required to solve and complete many of these structures because of the need for manual interpretation of complex numerical data using many software packages and the repeated use of interactive three-dimensional graphics. PHENIX has been developed to provide a comprehensive system for macromolecular crystallographic structure solution with an emphasis on the automation of all procedures. This has relied on the development of algorithms that minimize or eliminate subjective input, the development of algorithms that automate procedures that are traditionally performed by hand and, finally, the development of a framework that allows a tight integration between the algorithms. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_20267.map.gz emd_20267.map.gz | 141.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-20267-v30.xml emd-20267-v30.xml emd-20267.xml emd-20267.xml | 21.9 KB 21.9 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_20267.png emd_20267.png | 80.5 KB | ||
| Others |  emd_20267_half_map_1.map.gz emd_20267_half_map_1.map.gz emd_20267_half_map_2.map.gz emd_20267_half_map_2.map.gz | 139 MB 139 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-20267 http://ftp.pdbj.org/pub/emdb/structures/EMD-20267 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-20267 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-20267 | HTTPS FTP |
-Related structure data
| Related structure data |  6p7nMC  6p7mC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_20267.map.gz / Format: CCP4 / Size: 149.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_20267.map.gz / Format: CCP4 / Size: 149.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | 3D Reconstruction (State II) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.9 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Half map: 3D Reconstruction (State II) Half Map A
| File | emd_20267_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | 3D Reconstruction (State II) Half Map A | ||||||||||||
| Projections & Slices |
| ||||||||||||
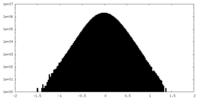
| Density Histograms |
-Half map: 3D Reconstruction (State II) Half Map B
| File | emd_20267_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | 3D Reconstruction (State II) Half Map B | ||||||||||||
| Projections & Slices |
| ||||||||||||
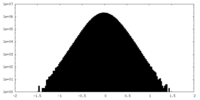
| Density Histograms |
- Sample components
Sample components
-Entire : LbCas12a-crRNA-AcrVA4 Complex (2:2, State I)
| Entire | Name: LbCas12a-crRNA-AcrVA4 Complex (2:2, State I) |
|---|---|
| Components |
|
-Supramolecule #1: LbCas12a-crRNA-AcrVA4 Complex (2:2, State I)
| Supramolecule | Name: LbCas12a-crRNA-AcrVA4 Complex (2:2, State I) / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#3 Details: Homodimeric AcrVA4 bound to two copies of the LbCas12a-crRNA complex. |
|---|
-Supramolecule #2: LbCas12a
| Supramolecule | Name: LbCas12a / type: complex / ID: 2 / Parent: 1 / Macromolecule list: #2 |
|---|---|
| Source (natural) | Organism:  Lachnospiraceae bacterium ND2006 (bacteria) Lachnospiraceae bacterium ND2006 (bacteria) |
-Supramolecule #3: crRNA
| Supramolecule | Name: crRNA / type: complex / ID: 3 / Parent: 1 / Macromolecule list: #3 |
|---|---|
| Source (natural) | Organism:  Lachnospiraceae bacterium ND2006 (bacteria) Lachnospiraceae bacterium ND2006 (bacteria) |
-Supramolecule #4: AcrVA4
| Supramolecule | Name: AcrVA4 / type: complex / ID: 4 / Parent: 1 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Moraxella bovoculi (bacteria) Moraxella bovoculi (bacteria) |
-Macromolecule #1: anti-CRISPR VA4
| Macromolecule | Name: anti-CRISPR VA4 / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Moraxella bovoculi (bacteria) Moraxella bovoculi (bacteria) |
| Molecular weight | Theoretical: 27.641422 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: SNAMYEIKLN DTLIHQTDDR VNAFVAYRYL LRRGDLPKCE NIARMYYDGK VIKTDVIDHD SVHSDEQAKV SNNDIIKMAI SELGVNNFK SLIKKQGYPF SNGHINSWFT DDPVKSKTMH NDEMYLVVQA LIRACIIKEI DLYTEQLYNI IKSLPYDKRP N VVYSDQPL ...String: SNAMYEIKLN DTLIHQTDDR VNAFVAYRYL LRRGDLPKCE NIARMYYDGK VIKTDVIDHD SVHSDEQAKV SNNDIIKMAI SELGVNNFK SLIKKQGYPF SNGHINSWFT DDPVKSKTMH NDEMYLVVQA LIRACIIKEI DLYTEQLYNI IKSLPYDKRP N VVYSDQPL DPNNLDLSEP ELWAEQVGEC MRYAHNDQPC FYIGSTKREL RVNYIVPVIG VRDEIERVMT LEEVRNLHK UniProtKB: Uncharacterized protein |
-Macromolecule #2: Cas12a
| Macromolecule | Name: Cas12a / type: protein_or_peptide / ID: 2 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Lachnospiraceae bacterium ND2006 (bacteria) Lachnospiraceae bacterium ND2006 (bacteria) |
| Molecular weight | Theoretical: 144.160609 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: SNAMSKLEKF TNCYSLSKTL RFKAIPVGKT QENIDNKRLL VEDEKRAEDY KGVKKLLDRY YLSFINDVLH SIKLKNLNNY ISLFRKKTR TEKENKELEN LEINLRKEIA KAFKGNEGYK SLFKKDIIET ILPEFLDDKD EIALVNSFNG FTTAFTGFFD N RENMFSEE ...String: SNAMSKLEKF TNCYSLSKTL RFKAIPVGKT QENIDNKRLL VEDEKRAEDY KGVKKLLDRY YLSFINDVLH SIKLKNLNNY ISLFRKKTR TEKENKELEN LEINLRKEIA KAFKGNEGYK SLFKKDIIET ILPEFLDDKD EIALVNSFNG FTTAFTGFFD N RENMFSEE AKSTSIAFRC INENLTRYIS NMDIFEKVDA IFDKHEVQEI KEKILNSDYD VEDFFEGEFF NFVLTQEGID VY NAIIGGF VTESGEKIKG LNEYINLYNQ KTKQKLPKFK PLYKQVLSDR ESLSFYGEGY TSDEEVLEVF RNTLNKNSEI FSS IKKLEK LFKNFDEYSS AGIFVKNGPA ISTISKDIFG EWNVIRDKWN AEYDDIHLKK KAVVTEKYED DRRKSFKKIG SFSL EQLQE YADADLSVVE KLKEIIIQKV DEIYKVYGSS EKLFDADFVL EKSLKKNDAV VAIMKDLLDS VKSFENYIKA FFGEG KETN RDESFYGDFV LAYDILLKVD HIYDAIRNYV TQKPYSKDKF KLYFQNPQFM GGWDKDKETD YRATILRYGS KYYLAI MDK KYAKCLQKID KDDVNGNYEK INYKLLPGPN KMLPKVFFSK KWMAYYNPSE DIQKIYKNGT FKKGDMFNLN DCHKLID FF KDSISRYPKW SNAYDFNFSE TEKYKDIAGF YREVEEQGYK VSFESASKKE VDKLVEEGKL YMFQIYNKDF SDKSHGTP N LHTMYFKLLF DENNHGQIRL SGGAELFMRR ASLKKEELVV HPANSPIANK NPDNPKKTTT LSYDVYKDKR FSEDQYELH IPIAINKCPK NIFKINTEVR VLLKHDDNPY VIGIDRGERN LLYIVVVDGK GNIVEQYSLN EIINNFNGIR IKTDYHSLLD KKEKERFEA RQNWTSIENI KELKAGYISQ VVHKICELVE KYDAVIALED LNSGFKNSRV KVEKQVYQKF EKMLIDKLNY M VDKKSNPC ATGGALKGYQ ITNKFESFKS MSTQNGFIFY IPAWLTSKID PSTGFVNLLK TKYTSIADSK KFISSFDRIM YV PEEDLFE FALDYKNFSR TDADYIKKWK LYSYGNRIRI FRNPKKNNVF DWEEVCLTSA YKELFNKYGI NYQQGDIRAL LCE QSDKAF YSSFMALMSL MLQMRNSITG RTDVDFLISP VKNSDGIFYD SRNYEAQENA ILPKNADANG AYNIARKVLW AIGQ FKKAE DEKLDKVKIA ISNKEWLEYA QTSVKH |
-Macromolecule #3: mature crRNA
| Macromolecule | Name: mature crRNA / type: rna / ID: 3 / Number of copies: 2 |
|---|---|
| Source (natural) | Organism:  Lachnospiraceae bacterium ND2006 (bacteria) Lachnospiraceae bacterium ND2006 (bacteria) |
| Molecular weight | Theoretical: 12.815634 KDa |
| Sequence | String: AAUUUCUACU AAGUGUAGAU AAAGUGCUCA UCAUUGGAAA |
-Macromolecule #4: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 4 / Number of copies: 4 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 Component:
| |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Grid | Details: unspecified | |||||||||||||||
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: SUPER-RESOLUTION / Average electron dose: 42.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: PDB ENTRY PDB model - PDB ID: |
|---|---|
| Final reconstruction | Number classes used: 1 / Applied symmetry - Point group: C1 (asymmetric) / Resolution.type: BY AUTHOR / Resolution: 4.9 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 79787 |
| Initial angle assignment | Type: NOT APPLICABLE |
| Final angle assignment | Type: NOT APPLICABLE |
 Movie
Movie Controller
Controller













 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)