[English] 日本語
 Yorodumi
Yorodumi- EMDB-0705: Structure of LbCas12a-crRNA complex bound to AcrVA4 (form B complex) -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-0705 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of LbCas12a-crRNA complex bound to AcrVA4 (form B complex) | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | CRISPR-Cas / anti-CRISPR / RNA BINDING PROTEIN / RNA BINDING PROTEIN-RNA complex | |||||||||
| Function / homology | Uncharacterized protein Function and homology information Function and homology information | |||||||||
| Biological species |  Lachnospiraceae bacterium (bacteria) / Lachnospiraceae bacterium (bacteria) /  Moraxella bovoculi (bacteria) / synthetic construct (others) Moraxella bovoculi (bacteria) / synthetic construct (others) | |||||||||
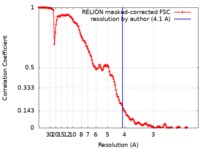
| Method | single particle reconstruction / cryo EM / Resolution: 4.1 Å | |||||||||
 Authors Authors | Peng R / Li Z | |||||||||
 Citation Citation |  Journal: Proc Natl Acad Sci U S A / Year: 2019 Journal: Proc Natl Acad Sci U S A / Year: 2019Title: Structural insight into multistage inhibition of CRISPR-Cas12a by AcrVA4. Authors: Ruchao Peng / Zhiteng Li / Ying Xu / Shaoshuai He / Qi Peng / Lian-Ao Wu / Ying Wu / Jianxun Qi / Peiyi Wang / Yi Shi / George F Gao /  Abstract: Prokaryotes possess CRISPR-Cas systems to exclude parasitic predators, such as phages and mobile genetic elements (MGEs). These predators, in turn, encode anti-CRISPR (Acr) proteins to evade the ...Prokaryotes possess CRISPR-Cas systems to exclude parasitic predators, such as phages and mobile genetic elements (MGEs). These predators, in turn, encode anti-CRISPR (Acr) proteins to evade the CRISPR-Cas immunity. Recently, AcrVA4, an Acr protein inhibiting the CRISPR-Cas12a system, was shown to diminish Cas12a (LbCas12a)-mediated genome editing in human cells, but the underlying mechanisms remain elusive. Here we report the cryo-EM structures of AcrVA4 bound to CRISPR RNA (crRNA)-loaded LbCas12a and found AcrVA4 could inhibit LbCas12a at several stages of the CRISPR-Cas working pathway, different from other characterized type I/II Acr inhibitors which target only 1 stage. First, it locks the conformation of the LbCas12a-crRNA complex to prevent target DNA-crRNA hybridization. Second, it interacts with the LbCas12a-crRNA-dsDNA complex to release the bound DNA before cleavage. Third, AcrVA4 binds the postcleavage LbCas12a complex to possibly block enzyme recycling. These findings highlight the multifunctionality of AcrVA4 and provide clues for developing regulatory genome-editing tools. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_0705.map.gz emd_0705.map.gz | 14.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-0705-v30.xml emd-0705-v30.xml emd-0705.xml emd-0705.xml | 16.3 KB 16.3 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_0705_fsc.xml emd_0705_fsc.xml | 14.2 KB | Display |  FSC data file FSC data file |
| Images |  emd_0705.png emd_0705.png | 55.8 KB | ||
| Filedesc metadata |  emd-0705.cif.gz emd-0705.cif.gz | 6.9 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-0705 http://ftp.pdbj.org/pub/emdb/structures/EMD-0705 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-0705 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-0705 | HTTPS FTP |
-Related structure data
| Related structure data |  6klbMC  0704C  6kl9C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_0705.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_0705.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.08 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : LbCas12a-crRNA complex bound to AcrVA4 (form B complex)
| Entire | Name: LbCas12a-crRNA complex bound to AcrVA4 (form B complex) |
|---|---|
| Components |
|
-Supramolecule #1: LbCas12a-crRNA complex bound to AcrVA4 (form B complex)
| Supramolecule | Name: LbCas12a-crRNA complex bound to AcrVA4 (form B complex) type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#3 |
|---|---|
| Source (natural) | Organism:  Lachnospiraceae bacterium (bacteria) Lachnospiraceae bacterium (bacteria) |
| Molecular weight | Theoretical: 300 KDa |
-Macromolecule #1: LbCas12a
| Macromolecule | Name: LbCas12a / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Lachnospiraceae bacterium (bacteria) Lachnospiraceae bacterium (bacteria) |
| Molecular weight | Theoretical: 143.888359 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MSKLEKFTNC YSLSKTLRFK AIPVGKTQEN IDNKRLLVED EKRAEDYKGV KKLLDRYYLS FINDVLHSIK LKNLNNYISL FRKKTRTEK ENKELENLEI NLRKEIAKAF KGNEGYKSLF KKDIIETILP EFLDDKDEIA LVNSFNGFTT AFTGFFDNRE N MFSEEAKS ...String: MSKLEKFTNC YSLSKTLRFK AIPVGKTQEN IDNKRLLVED EKRAEDYKGV KKLLDRYYLS FINDVLHSIK LKNLNNYISL FRKKTRTEK ENKELENLEI NLRKEIAKAF KGNEGYKSLF KKDIIETILP EFLDDKDEIA LVNSFNGFTT AFTGFFDNRE N MFSEEAKS TSIAFRCINE NLTRYISNMD IFEKVDAIFD KHEVQEIKEK ILNSDYDVED FFEGEFFNFV LTQEGIDVYN AI IGGFVTE SGEKIKGLNE YINLYNQKTK QKLPKFKPLY KQVLSDRESL SFYGEGYTSD EEVLEVFRNT LNKNSEIFSS IKK LEKLFK NFDEYSSAGI FVKNGPAIST ISKDIFGEWN VIRDKWNAEY DDIHLKKKAV VTEKYEDDRR KSFKKIGSFS LEQL QEYAD ADLSVVEKLK EIIIQKVDEI YKVYGSSEKL FDADFVLEKS LKKNDAVVAI MKDLLDSVKS FENYIKAFFG EGKET NRDE SFYGDFVLAY DILLKVDHIY DAIRNYVTQK PYSKDKFKLY FQNPQFMGGW DKDKETDYRA TILRYGSKYY LAIMDK KYA KCLQKIDKDD VNGNYEKINY KLLPGPNKML PKVFFSKKWM AYYNPSEDIQ KIYKNGTFKK GDMFNLNDCH KLIDFFK DS ISRYPKWSNA YDFNFSETEK YKDIAGFYRE VEEQGYKVSF ESASKKEVDK LVEEGKLYMF QIYNKDFSDK SHGTPNLH T MYFKLLFDEN NHGQIRLSGG AELFMRRASL KKEELVVHPA NSPIANKNPD NPKKTTTLSY DVYKDKRFSE DQYELHIPI AINKCPKNIF KINTEVRVLL KHDDNPYVIG IDRGERNLLY IVVVDGKGNI VEQYSLNEII NNFNGIRIKT DYHSLLDKKE KERFEARQN WTSIENIKEL KAGYISQVVH KICELVEKYD AVIALEDLNS GFKNSRVKVE KQVYQKFEKM LIDKLNYMVD K KSNPCATG GALKGYQITN KFESFKSMST QNGFIFYIPA WLTSKIDPST GFVNLLKTKY TSIADSKKFI SSFDRIMYVP EE DLFEFAL DYKNFSRTDA DYIKKWKLYS YGNRIRIFRN PKKNNVFDWE EVCLTSAYKE LFNKYGINYQ QGDIRALLCE QSD KAFYSS FMALMSLMLQ MRNSITGRTD VDFLISPVKN SDGIFYDSRN YEAQENAILP KNADANGAYN IARKVLWAIG QFKK AEDEK LDKVKIAISN KEWLEYAQTS VKH |
-Macromolecule #2: AcrVA4
| Macromolecule | Name: AcrVA4 / type: protein_or_peptide / ID: 2 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Moraxella bovoculi (bacteria) Moraxella bovoculi (bacteria) |
| Molecular weight | Theoretical: 27.369162 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MYEIKLNDTL IHQTDDRVNA FVAYRYLLRR GDLPKCENIA RMYYDGKVIK TDVIDHDSVH SDEQAKVSNN DIIKMAISEL GVNNFKSLI KKQGYPFSNG HINSWFTDDP VKSKTMHNDE MYLVVQALIR ACIIKEIDLY TEQLYNIIKS LPYDKRPNVV Y SDQPLDPN ...String: MYEIKLNDTL IHQTDDRVNA FVAYRYLLRR GDLPKCENIA RMYYDGKVIK TDVIDHDSVH SDEQAKVSNN DIIKMAISEL GVNNFKSLI KKQGYPFSNG HINSWFTDDP VKSKTMHNDE MYLVVQALIR ACIIKEIDLY TEQLYNIIKS LPYDKRPNVV Y SDQPLDPN NLDLSEPELW AEQVGECMRY AHNDQPCFYI GSTKRELRVN YIVPVIGVRD EIERVMTLEE VRNLHK UniProtKB: Uncharacterized protein |
-Macromolecule #3: crRNA
| Macromolecule | Name: crRNA / type: rna / ID: 3 / Number of copies: 2 |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 13.48202 KDa |
| Sequence | String: AAUUUCUACU AAGUGUAGAU CGGUCUCGCA AAGAAUGGAU AU |
-Macromolecule #4: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 4 / Number of copies: 4 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
-Macromolecule #5: water
| Macromolecule | Name: water / type: ligand / ID: 5 / Number of copies: 11 / Formula: HOH |
|---|---|
| Molecular weight | Theoretical: 18.015 Da |
| Chemical component information |  ChemComp-HOH: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1.0 mg/mL |
|---|---|
| Buffer | pH: 7.5 |
| Grid | Model: Homemade / Material: COPPER / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 45 sec. / Pretreatment - Atmosphere: AIR |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Bioquantum / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: SUPER-RESOLUTION / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller











 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)