[English] 日本語
 Yorodumi
Yorodumi- EMDB-26574: KS-AT di-domain of mycobacterial Pks13 with endogenous KS ligand bound -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | KS-AT di-domain of mycobacterial Pks13 with endogenous KS ligand bound | ||||||||||||
 Map data Map data | |||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | mycolic acid synthesis / ketosynthase / acyltransferase / multi-domain assembly / BIOSYNTHETIC PROTEIN | ||||||||||||
| Function / homology |  Function and homology information Function and homology information6-deoxyerythronolide-B synthase / erythronolide synthase activity / DIM/DIP cell wall layer assembly / fatty acid synthase activity / phosphopantetheine binding / fatty acid biosynthetic process / plasma membrane / cytoplasm Similarity search - Function | ||||||||||||
| Biological species |  Mycolicibacterium smegmatis MC2 155 (bacteria) Mycolicibacterium smegmatis MC2 155 (bacteria) | ||||||||||||
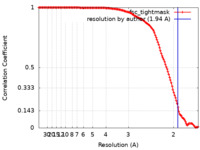
| Method | single particle reconstruction / cryo EM / Resolution: 1.94 Å | ||||||||||||
 Authors Authors | Kim SK / Dickinson MS / Finer-Moore JS / Rosenberg OS / Stroud RM | ||||||||||||
| Funding support |  United States, 3 items United States, 3 items
| ||||||||||||
 Citation Citation |  Journal: Nat Struct Mol Biol / Year: 2023 Journal: Nat Struct Mol Biol / Year: 2023Title: Structure and dynamics of the essential endogenous mycobacterial polyketide synthase Pks13. Authors: Sun Kyung Kim / Miles Sasha Dickinson / Janet Finer-Moore / Ziqiang Guan / Robyn M Kaake / Ignacia Echeverria / Jen Chen / Ernst H Pulido / Andrej Sali / Nevan J Krogan / Oren S Rosenberg / Robert M Stroud /  Abstract: The mycolic acid layer of the Mycobacterium tuberculosis cell wall is essential for viability and virulence, and the enzymes responsible for its synthesis are targets for antimycobacterial drug ...The mycolic acid layer of the Mycobacterium tuberculosis cell wall is essential for viability and virulence, and the enzymes responsible for its synthesis are targets for antimycobacterial drug development. Polyketide synthase 13 (Pks13) is a module encoding several enzymatic and transport functions that carries out the condensation of two different long-chain fatty acids to produce mycolic acids. We determined structures by cryogenic-electron microscopy of dimeric multi-enzyme Pks13 purified from mycobacteria under normal growth conditions, captured with native substrates. Structures define the ketosynthase (KS), linker and acyl transferase (AT) domains at 1.8 Å resolution and two alternative locations of the N-terminal acyl carrier protein. These structures suggest intermediate states on the pathway for substrate delivery to the KS domain. Other domains, visible at lower resolution, are flexible relative to the KS-AT core. The chemical structures of three bound endogenous long-chain fatty acid substrates were determined by electrospray ionization mass spectrometry. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_26574.map.gz emd_26574.map.gz | 108.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-26574-v30.xml emd-26574-v30.xml emd-26574.xml emd-26574.xml | 27.3 KB 27.3 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_26574_fsc.xml emd_26574_fsc.xml | 14.5 KB | Display |  FSC data file FSC data file |
| Images |  emd_26574.png emd_26574.png | 74.6 KB | ||
| Filedesc metadata |  emd-26574.cif.gz emd-26574.cif.gz | 7.7 KB | ||
| Others |  emd_26574_additional_1.map.gz emd_26574_additional_1.map.gz emd_26574_additional_2.map.gz emd_26574_additional_2.map.gz emd_26574_half_map_1.map.gz emd_26574_half_map_1.map.gz emd_26574_half_map_2.map.gz emd_26574_half_map_2.map.gz | 14 MB 185.7 MB 202.8 MB 202.8 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-26574 http://ftp.pdbj.org/pub/emdb/structures/EMD-26574 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-26574 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-26574 | HTTPS FTP |
-Validation report
| Summary document |  emd_26574_validation.pdf.gz emd_26574_validation.pdf.gz | 685.6 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_26574_full_validation.pdf.gz emd_26574_full_validation.pdf.gz | 685.2 KB | Display | |
| Data in XML |  emd_26574_validation.xml.gz emd_26574_validation.xml.gz | 21.7 KB | Display | |
| Data in CIF |  emd_26574_validation.cif.gz emd_26574_validation.cif.gz | 28 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-26574 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-26574 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-26574 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-26574 | HTTPS FTP |
-Related structure data
| Related structure data |  7uk4MC  8cuyC  8cuzC  8cv0C  8cv1C C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_26574.map.gz / Format: CCP4 / Size: 219.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_26574.map.gz / Format: CCP4 / Size: 219.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.835 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: density-modified, sharpened map
| File | emd_26574_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | density-modified, sharpened map | ||||||||||||
| Projections & Slices |
| ||||||||||||
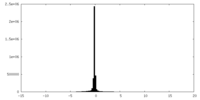
| Density Histograms |
-Additional map: DeepEMhancer sharpened map contour at 0.268
| File | emd_26574_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | DeepEMhancer sharpened map contour at 0.268 | ||||||||||||
| Projections & Slices |
| ||||||||||||
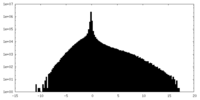
| Density Histograms |
-Half map: #2
| File | emd_26574_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
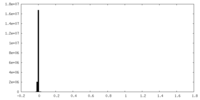
| Density Histograms |
-Half map: #1
| File | emd_26574_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
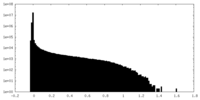
| Density Histograms |
- Sample components
Sample components
-Entire : Mycobacterial polyketide synthase 13
| Entire | Name: Mycobacterial polyketide synthase 13 |
|---|---|
| Components |
|
-Supramolecule #1: Mycobacterial polyketide synthase 13
| Supramolecule | Name: Mycobacterial polyketide synthase 13 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 Details: The gene for Mycobacterium smegmatis polyketide synthase 13 (Pks13) was tagged with TEV-cleavable GFP at its C-terminus and purified from its natural source with anti-GFP nanobody beads. GFP ...Details: The gene for Mycobacterium smegmatis polyketide synthase 13 (Pks13) was tagged with TEV-cleavable GFP at its C-terminus and purified from its natural source with anti-GFP nanobody beads. GFP was cleaved to yield the full-length Pks13. |
|---|---|
| Source (natural) | Organism:  Mycolicibacterium smegmatis MC2 155 (bacteria) Mycolicibacterium smegmatis MC2 155 (bacteria) |
-Supramolecule #2: Ketosynthase domain
| Supramolecule | Name: Ketosynthase domain / type: complex / ID: 2 / Parent: 1 / Macromolecule list: #1 Details: Linked to acyl transferase domain at the C-terminus |
|---|---|
| Source (natural) | Organism:  Mycolicibacterium smegmatis MC2 155 (bacteria) Mycolicibacterium smegmatis MC2 155 (bacteria) |
-Supramolecule #3: Acyl transferase domain
| Supramolecule | Name: Acyl transferase domain / type: complex / ID: 3 / Parent: 1 / Macromolecule list: #1 / Details: linked to ketosynthase domain at the N-terminus |
|---|---|
| Source (natural) | Organism:  Mycolicibacterium smegmatis MC2 155 (bacteria) Mycolicibacterium smegmatis MC2 155 (bacteria) |
-Macromolecule #1: Polyketide synthase PKS13
| Macromolecule | Name: Polyketide synthase PKS13 / type: protein_or_peptide / ID: 1 Details: The gene for M. smegmatis polyketide synthase 13 (Pks13) was tagged with TEV-cleavable GFP at its C-terminus and purified from its natural source with anti-GFP nanobody beads. GFP was ...Details: The gene for M. smegmatis polyketide synthase 13 (Pks13) was tagged with TEV-cleavable GFP at its C-terminus and purified from its natural source with anti-GFP nanobody beads. GFP was cleaved to yield the full-length Pks13. Number of copies: 2 / Enantiomer: LEVO / EC number: 6-deoxyerythronolide-B synthase |
|---|---|
| Source (natural) | Organism:  Mycolicibacterium smegmatis MC2 155 (bacteria) / Strain: ATCC 700084 / mc(2)155 Mycolicibacterium smegmatis MC2 155 (bacteria) / Strain: ATCC 700084 / mc(2)155 |
| Molecular weight | Theoretical: 198.633359 KDa |
| Sequence | String: MDENHSDSNL PEGTEPAGRP APRTDMTVNE MREWLRNWVA NATGQSADAI DESTPMVELG LSSRDAVAMA SDIEDLTGVT LTATVAFRH PTIESLATVI IEGEPEPEPY DEDEDWSRTR DVEDIAIVGV ATRFPGDLNT PDEMWEALLE GKDCVTDLPE D RWTEFLDE ...String: MDENHSDSNL PEGTEPAGRP APRTDMTVNE MREWLRNWVA NATGQSADAI DESTPMVELG LSSRDAVAMA SDIEDLTGVT LTATVAFRH PTIESLATVI IEGEPEPEPY DEDEDWSRTR DVEDIAIVGV ATRFPGDLNT PDEMWEALLE GKDCVTDLPE D RWTEFLDE PRIAERVKKA RTRGGYLTDI KGFDSEFFAL SKMEADNIDP QQRMALELTW EALEHARIPA SSLRGESVGV YI GSSTNDY SFLAMSDPSI AHPYAITGTA SSIIANRVSY FYDFRGPSVA VDTACSSSLV ATHQGVQALR AGEADVAIVG GVN ALVTPL VTVGFDEVGG VLAPDGRIKS FSSDADGYAR SEGGGMLVLK RISDARRDGD QILAVIAGSA VNHDGRSNGL LAPN PDAQA EVLRKAYKDA GINPRDVDYI EAHGTGTILG DPIEADALGR IVGKGRPADK PALLGAVKSN LGHLESAAGA ASLAK MTLA LANDKLPPSI NYAGPNPYID FEKERLKVND TVSDWPRYSG KAIAGVSGFG FGGANAHVVM REVLAGDLVE PEPEPE PEA KPEKSEADAV YVGGVRMDEY GEFIDEDEPA EGGDAYPSYD EDSYELPGIT EAAQRLLEQA REELEAKEAE EPTKQLV PL AVSAFLTSRK RQAAAELADW IDSPEGRASS LESIGRSLSR RNHGRSRAVV LAHDHDEAIK GLRALAEGKQ HPSVLSAD G PVTNGPVWVL AGFGAQHRKM GKSLYLRNEV FAEWINKVDA LIQDERGYSI LELILDDNVD YTDATCEYPI EVVQLVIFA IQIALGELLR HHGAKPAAVV GQSLGEAAAS YFAGGLSLAD ATRTICSRSH LMGEGEAMLF GEYIRLMALV EYSADEIKTV FSDYPDLEV CVYAAPTQTV IGGPPDQVDA IIARAESEGK FARKFQTKGA SHTQQMDPLL GELAAELQGI EPKPLTTGYF S TVHEGTFI RPGSAPIHDV DYWKKGLRHS VYFTQGIRNA VDNGHTTFLE LAPNPVALMQ VGLTTASAGL HDAQLIATLA RK QDEVESM ISAMAQLYVH GHDLDFRTLF PRRSKGLAGA LDFANIPPTR FKRKEHWLPA HFTGDSSAVM PGNHVATPDG RHV WEFVPR GKTDLAALVK AAAAQVLPDA KLAAFEQRAV PADNARLVTT LTRHPGGATV QVHARVEESF TLVYDAIVAR ANGA GVTAL PVAVGAGVAV SGDVAGEGAG ASVIEDDEPD AEILQDNLTA GAGMGADFQK WDPNSGETIG QRLGTIVGAA MGYEP EDLP WEVPLIELGL DSLMAVRIKN RVEYDFDLPP IQLTAVRDAN LYNVEELIRY AIEHRDEVEQ IAESQKGKTA EEIAAE QSE LLGGASTVAE LEAKLAEAGH PLAAKDSEDS ENSEDNAAGA AAAAEASAVE GLEIPPPPTD PTGPGGAPIP PPPSDPS GP AQAASATDAP AGTVNKATAA AAAAKVLTQE AVTEALGADV PPRDAAERVT FATWAIVTGK SPGGIFNELP TVSEETAK K MAERLSERAE GTITVEDVLG AKTIEGLATI VREQLEEGVV DGFVRTLRPP KEGSNAVPLF VFHPAGGSTV VYEPLMKRL PADVPVYGLE RVEGSIEERA AEYVPKLLEM HKGPFVLAGW SLGGALAYAC AIGLKQSGAD VRFVGLIDTV LPGEPIDQSK EGMRARWDR YARFAERTFN VEIPAIPYEE LEKLDDEGQV KYVLEIVKES GVQIPGGIIE HQRTSYLDNR ALDTVDIKPY D GHVTLYMA DRYHDDAIVF EPAYATRKPD GGWGSFVSDL EVVHIGGEHI QAIDEPYIAK VGAHMSEALN RIEAQASKED GA KSKSTSE NLYFQ UniProtKB: Polyketide synthase PKS13 |
-Macromolecule #2: UNKNOWN LIGAND
| Macromolecule | Name: UNKNOWN LIGAND / type: ligand / ID: 2 Details: The fatty acid ligand designated as UNL (unknown ligand) is a divided population of fatty acids of formula C55H106O2 and C40H78O2 as confirmed by mass spectrometry. Number of copies: 2 / Formula: UNL |
|---|---|
| Molecular weight | Theoretical: 312.53 Da |
| Chemical component information | 
ChemComp-UNL: |
-Macromolecule #3: water
| Macromolecule | Name: water / type: ligand / ID: 3 / Number of copies: 390 / Formula: HOH |
|---|---|
| Molecular weight | Theoretical: 18.015 Da |
| Chemical component information |  ChemComp-HOH: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1.5 mg/mL | ||||||
|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 / Component:
| ||||||
| Grid | Model: Quantifoil R1.2/1.3 / Material: GOLD / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE | ||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 283.15 K / Instrument: FEI VITROBOT MARK IV Details: Full length pks13 was concentrated to 1.5 mg mL-1 for cryo-EM grid preparation. 4.5 uL of sample was applied to freshly glow discharged holey carbon on gold R1.2/1.3 300 mesh Quantifoil ...Details: Full length pks13 was concentrated to 1.5 mg mL-1 for cryo-EM grid preparation. 4.5 uL of sample was applied to freshly glow discharged holey carbon on gold R1.2/1.3 300 mesh Quantifoil grids and blotted for 9 s with Whatman 1 filter paper at max humidity and 10oC in a FEI Mark IV Vitrobot, before vitrification in liquid nitrogen-cooled liquid ethane.. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Number real images: 7567 / Average exposure time: 5.9 sec. / Average electron dose: 45.8 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 70.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 1.5 µm / Nominal defocus min: 0.7000000000000001 µm / Nominal magnification: 105000 |
| Sample stage | Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: AB INITIO MODEL |
|---|---|
| Output model |  PDB-7uk4: |
 Movie
Movie Controller
Controller










 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)