-Search query
-Search result
Showing 1 - 50 of 102 items for (author: muench & s)

EMDB-17725: 
CryoEM reconstruction of Influenza A virus (HK68) hemagglutinin bound to an Affimer reagent

EMDB-36800: 
Potassium transporter KtrAB from Bacillus subtilis in ADP-bound state

EMDB-36801: 
Potassium transporter KtrAB from Bacillus subtilis in ADP-bound state, focused refined on KtrA octamer

EMDB-36802: 
Potassium transporter KtrAB from Bacillus subtilis in ADP-bound state, focused refined on KtrB dimer

EMDB-36803: 
Potassium transporter KtrAB from Bacillus subtilis in ATP-bound state with addition of MgCl2

EMDB-36804: 
Potassium transporter KtrAB from Bacillus subtilis in ATP-bound state with addition of EDTA and EGTA

EMDB-38477: 
Potassium transporter KtrAB from Bacillus subtilis in ATP-bound state with addition of EDTA and EGTA, vertical C2 symmetry axis

EMDB-38478: 
Potassium transporter KtrAB from Bacillus subtilis in ATP-bound state with addition of EDTA and EGTA, C1 symmetry

PDB-8k1s: 
Potassium transporter KtrAB from Bacillus subtilis in ADP-bound state

PDB-8k1t: 
Potassium transporter KtrAB from Bacillus subtilis in ATP-bound state with addition of MgCl2

PDB-8k1u: 
Potassium transporter KtrAB from Bacillus subtilis in ATP-bound state with addition of EDTA and EGTA

PDB-8xmh: 
Potassium transporter KtrAB from Bacillus subtilis in ATP-bound state with addition of EDTA and EGTA, vertical C2 symmetry axis

PDB-8xmi: 
Potassium transporter KtrAB from Bacillus subtilis in ATP-bound state with addition of EDTA and EGTA, C1 symmetry
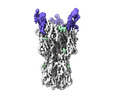
EMDB-17724: 
CryoEM reconstruction of hemagglutinin HK68 of Influenza A virus bound to an Affimer reagent

PDB-8pk3: 
CryoEM reconstruction of hemagglutinin HK68 of Influenza A virus bound to an Affimer reagent

EMDB-16041: 
CryoEM structure of quinol-dependent Nitric Oxide Reductase (qNOR) from Alcaligenes xylosoxidans at 2.2 A resolution

PDB-8bgw: 
CryoEM structure of quinol-dependent Nitric Oxide Reductase (qNOR) from Alcaligenes xylosoxidans at 2.2 A resolution

EMDB-14981: 
Structure of b-galactosidase without reducing agent prepared by Vitrobot

EMDB-14982: 
Structure of b-galactosidase with DTT prepared by Vitrobot

EMDB-14983: 
Structure of b-galactosidase with TCEP prepared by Vitrobot

EMDB-14984: 
Structure of b-galactosidase on continuous carbon support film prepared by Vitrobot

EMDB-15162: 
Structure of the human mitochondrial HSPD1 single ring from grids produced in 14ms

EMDB-15167: 
Structure of the human mitochondrial HSPD1 single ring in the presence of Methionine

EMDB-15176: 
Structure of the human mitochondrial HSPD1 single ring in the presence of ascorbate

EMDB-15178: 
Structure of the human mitochondrial HSPD1 single ring (blotted but no DTT)

EMDB-13421: 
Cryo-EM maps of the RET/GDF15/GFRAL complex

EMDB-13688: 
Negative stain EM map of the extracellular domain of the RET(C634R) mutant dimer
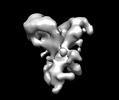
EMDB-13689: 
Negative stain EM map of the extracellular domain of the RET(C634R)/GDF15/GFRAL complex

EMDB-12043: 
AcrB in cycloalkane amphipol

PDB-7b5p: 
AcrB in cycloalkane amphipol

EMDB-13328: 
Time-resolved cryo-EM structures of ATP-induced actomyosin dissociation

EMDB-11715: 
Cryo-EM reconstruction of the di-hexameric endocytic adaptor complex AENTH (ENTH F5A/L12A/V13A mutant)

EMDB-11987: 
Structure of the endocytic adaptor complex AENTH

PDB-7b2l: 
Structure of the endocytic adaptor complex AENTH

EMDB-11189: 
Structure of human mitochondrial HSPD1 as an inverted double ring

EMDB-11950: 
Structure of the human mitochondrial HSPD1 single ring

PDB-7azp: 
Structure of the human mitochondrial HSPD1 single ring

EMDB-10903: 
Human TRPC5 in complex with Pico145 (HC-608)

EMDB-10909: 
Human TRPC5 in the presence of 50 uM Pico145 (HC-608)

EMDB-10910: 
Human TRPC5 in the presence of 20 uM ZnCl2

PDB-6ysn: 
Human TRPC5 in complex with Pico145 (HC-608)

EMDB-10871: 
30S ribosome subunit deposited by spraying (13 ms delay)

EMDB-10872: 
30S ribosome subunit deposited using the chameleon (54 ms delay)

EMDB-10873: 
30S ribosome subunit deposited using the chameleon (200 ms delay)

EMDB-10874: 
30S ribosome subunit prepared by blotting

EMDB-10875: 
50S ribosome subunit deposited by spraying (13 ms delay)

EMDB-10876: 
50S ribosome subunit deposited using the chameleon (54 ms delay)

EMDB-10877: 
50S ribosome subunit deposited using the chameleon (200 ms delay)

EMDB-10878: 
50S ribosome subunit prepared by blotting

EMDB-10879: 
70S ribosome deposited by spraying (13 ms delay)
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
