+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM maps of the RET/GDF15/GFRAL complex | ||||||||||||||||||
 Map data Map data | Post-processed masked map. Map has been z-flipped to match the orientation of the published structure of the RET/GDF15/GRFAL complex (PDB ID 6Q2J). | ||||||||||||||||||
 Sample Sample |
| ||||||||||||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | ||||||||||||||||||
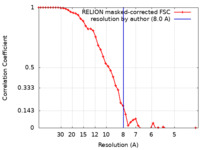
| Method | single particle reconstruction / cryo EM / Resolution: 8.0 Å | ||||||||||||||||||
 Authors Authors | Liu YL / Muench SP / Goldman A | ||||||||||||||||||
| Funding support |  United Kingdom, 5 items United Kingdom, 5 items
| ||||||||||||||||||
 Citation Citation |  Journal: J Biol Chem / Year: 2022 Journal: J Biol Chem / Year: 2022Title: Unexpected structures formed by the kinase RET C634R mutant extracellular domain suggest potential oncogenic mechanisms in MEN2A. Authors: Yixin Liu / Orquidea De Castro Ribeiro / Outi Haapanen / Gregory B Craven / Vivek Sharma / Stephen P Muench / Adrian Goldman /   Abstract: The RET receptor tyrosine kinase plays a pivotal role in cell survival, proliferation, and differentiation, and its abnormal activation leads to cancers through receptor fusions or point mutations. ...The RET receptor tyrosine kinase plays a pivotal role in cell survival, proliferation, and differentiation, and its abnormal activation leads to cancers through receptor fusions or point mutations. Mutations that disrupt the disulfide network in the extracellular domain (ECD) of RET drive multiple endocrine neoplasia type 2A (MEN2A), a hereditary syndrome associated with the development of thyroid cancers. However, structural details of how specific mutations affect RET are unclear. Here, we present the first structural insights into the ECD of the RET(C634R) mutant, the most common mutation in MEN2A. Using electron microscopy, we demonstrate that the C634R mutation causes ligand-independent dimerization of the RET ECD, revealing an unusual tail-to-tail conformation that is distinct from the ligand-induced signaling dimer of WT RET. Additionally, we show that the RET ECD dimer can form complexes with at least two of the canonical RET ligands and that these complexes form very different structures than WT RET ECD upon ligand binding. In conclusion, this structural analysis of cysteine-mutant RET ECD suggests a potential key mechanism of cancer induction in MEN2A, both in the absence and presence of its native ligands, and may offer new targets for therapeutic intervention. | ||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_13421.map.gz emd_13421.map.gz | 1.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-13421-v30.xml emd-13421-v30.xml emd-13421.xml emd-13421.xml | 24.7 KB 24.7 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_13421_fsc.xml emd_13421_fsc.xml | 4.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_13421.png emd_13421.png | 67.7 KB | ||
| Masks |  emd_13421_msk_1.map emd_13421_msk_1.map | 8 MB |  Mask map Mask map | |
| Others |  emd_13421_additional_1.map.gz emd_13421_additional_1.map.gz emd_13421_additional_2.map.gz emd_13421_additional_2.map.gz emd_13421_additional_3.map.gz emd_13421_additional_3.map.gz emd_13421_half_map_1.map.gz emd_13421_half_map_1.map.gz emd_13421_half_map_2.map.gz emd_13421_half_map_2.map.gz | 1.4 MB 1.3 MB 1.3 MB 5.4 MB 5.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-13421 http://ftp.pdbj.org/pub/emdb/structures/EMD-13421 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-13421 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-13421 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_13421.map.gz / Format: CCP4 / Size: 8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_13421.map.gz / Format: CCP4 / Size: 8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Post-processed masked map. Map has been z-flipped to match the orientation of the published structure of the RET/GDF15/GRFAL complex (PDB ID 6Q2J). | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2.13 Å | ||||||||||||||||||||||||||||||||||||
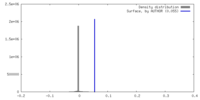
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_13421_msk_1.map emd_13421_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
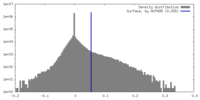
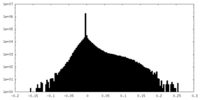
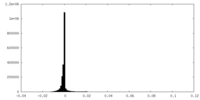
| Density Histograms |
-Additional map: Different conformations of the RET/GDF15/GFRAL complex (Class 2)
| File | emd_13421_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Different conformations of the RET/GDF15/GFRAL complex (Class 2) | ||||||||||||
| Projections & Slices |
| ||||||||||||
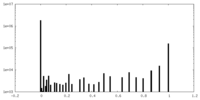
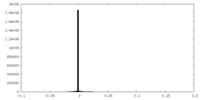
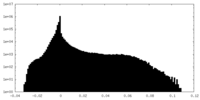
| Density Histograms |
-Additional map: Different conformations of the RET/GDF15/GFRAL complex (Class 1)
| File | emd_13421_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Different conformations of the RET/GDF15/GFRAL complex (Class 1) | ||||||||||||
| Projections & Slices |
| ||||||||||||
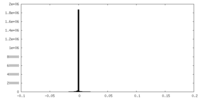
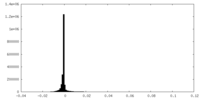
| Density Histograms |
-Additional map: Different conformations of the RET/GDF15/GFRAL complex (Class 3)
| File | emd_13421_additional_3.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Different conformations of the RET/GDF15/GFRAL complex (Class 3) | ||||||||||||
| Projections & Slices |
| ||||||||||||
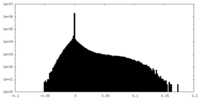
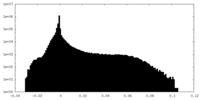
| Density Histograms |
-Half map: Half map2, z-flipped
| File | emd_13421_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map2, z-flipped | ||||||||||||
| Projections & Slices |
| ||||||||||||
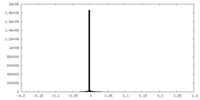
| Density Histograms |
-Half map: Half map1, z-flipped
| File | emd_13421_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map1, z-flipped | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : The extracellular domain complex of RET/GDF15/GFRAL
| Entire | Name: The extracellular domain complex of RET/GDF15/GFRAL |
|---|---|
| Components |
|
-Supramolecule #1: The extracellular domain complex of RET/GDF15/GFRAL
| Supramolecule | Name: The extracellular domain complex of RET/GDF15/GFRAL / type: complex / ID: 1 / Parent: 0 Details: The extracellular domain of the proto-oncogene tyrosine-protein kinase receptor Ret. Mature growth and differentiation factor 15 with two N-terminal Fc tags. The extracellular domain of GDNF ...Details: The extracellular domain of the proto-oncogene tyrosine-protein kinase receptor Ret. Mature growth and differentiation factor 15 with two N-terminal Fc tags. The extracellular domain of GDNF family receptor alpha-like. |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Experimental: 40 KDa |
-Supramolecule #2: The extracellular domain of proto-oncogene tyrosine-protein kinas...
| Supramolecule | Name: The extracellular domain of proto-oncogene tyrosine-protein kinase receptor Ret type: complex / ID: 2 / Parent: 1 Details: RET is secreted and purified from cell culture medium. |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Supramolecule #3: The mature growth and differentiation factor 15 with N-terminal F...
| Supramolecule | Name: The mature growth and differentiation factor 15 with N-terminal Fc tags. type: complex / ID: 3 / Parent: 1 Details: Fc-GDF15 was secreted and purified from cell culture medium using protein A resin. |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Supramolecule #4: The extracellular domain of GDNF family receptor alpha-like
| Supramolecule | Name: The extracellular domain of GDNF family receptor alpha-like type: complex / ID: 4 / Parent: 1 Details: GFRAL is secreted and purified from cell culture medium using Strep-Tactin affinity purification. |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.1 mg/mL | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 8 Component:
Details: Buffer was prepared fresh and filter steriled. | ||||||||||||
| Grid | Model: Quantifoil R1.2/1.3 / Material: GOLD / Mesh: 300 / Pretreatment - Type: GLOW DISCHARGE | ||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 80 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV / Details: Blot for 6 second before plunging. | ||||||||||||
| Details | This sample was >90% pure. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Detector mode: INTEGRATING / Number grids imaged: 1 / Average exposure time: 1.5 sec. / Average electron dose: 60.3 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 3.0 µm / Nominal defocus min: 1.6 µm / Nominal magnification: 75000 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller






 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)