-Search query
-Search result
Showing 1 - 50 of 233 items for (author: moore & jp)

EMDB-47765: 
Week 26 C3V5, gp41-GH and gp41-base epitope polyclonal antibodies from participant 202 in complex with ConM SOSIP
Method: single particle / : Lin RN, Torres JL, Tran AS, Ozorowski G, Ward AB

EMDB-70475: 
HIV-1 Env BG505 SOSIP.664-His in complex with PGT122 and 3BNC117 Fabs
Method: single particle / : Andrade TG, Ozorowski G, Ward AB

EMDB-70476: 
HIV-1 Env BG505 SOSIP.664-dPG-His in complex with PGT122 and 3BNC117 Fabs
Method: single particle / : Andrade TG, Ozorowski G, Ward AB

EMDB-48283: 
61-12A01 Fab in complex with HIV-1 GT1.1 v4.1 SOSIP Env trimer and RM20A3 Fab
Method: single particle / : Phulera S, Ozorowski G, Ward AB

EMDB-48286: 
206-3G08 Fab in complex with HIV-1 GT1.1 v4.1 SOSIP Env trimer and RM20A3 Fab
Method: single particle / : Phulera S, Ozorowski G, Ward AB

EMDB-48287: 
206-9C09 Fab in complex with HIV-1 GT1.1 v4.1 SOSIP Env trimer and RM20A3 Fab
Method: single particle / : Phulera S, Ozorowski G, Ward AB

EMDB-48290: 
273-4D01 Fab in complex with HIV-1 BG505 SOSIP Env trimer and RM20A3 Fab
Method: single particle / : Phulera S, Ozorowski G, Ward AB

EMDB-48291: 
253-7A03 Fab in complex with HIV-1 BG505 SOSIP Env trimer and RM20A3 Fab
Method: single particle / : Phulera S, Ozorowski G, Ward AB

EMDB-70490: 
BG505 GT1.1 SOSIP in complex with gp41-base epitope polyclonal antibodies isolated from a participant in the IAVI C101 clinical trial
Method: single particle / : Ozorowski G, Ward AB

EMDB-70491: 
BG505 GT1.1 SOSIP in complex with V1V2V3 epitope polyclonal antibodies isolated from a participant in the IAVI C101 clinical trial
Method: single particle / : Ozorowski G, Ward AB

EMDB-70492: 
BG505 GT1.1 SOSIP in complex with C3V5 epitope polyclonal antibodies isolated from a participant in the IAVI C101 clinical trial
Method: single particle / : Ozorowski G, Ward AB

EMDB-70493: 
BG505 GT1.1 SOSIP in complex with CD4bs epitope polyclonal antibodies isolated from a participant in the IAVI C101 clinical trial
Method: single particle / : Ozorowski G, Ward AB

EMDB-70494: 
BG505 GT1.1 SOSIP in complex with gp41 glycan hole epitope polyclonal antibodies isolated from a participant in the IAVI C101 clinical trial
Method: single particle / : Ozorowski G, Ward AB

EMDB-70495: 
BG505 GT1.1 SOSIP in complex with gp41 fusion peptide epitope polyclonal antibodies isolated from a participant in the IAVI C101 clinical trial
Method: single particle / : Ozorowski G, Ward AB

EMDB-41869: 
BG505.664 SOSIP in complex with polyclonal antibodies from NHP 8131 (gp120 glycan hole, gp41 glycan hole/fusion peptide and trimer base epitopes)
Method: single particle / : Zhang S, Ward AB

EMDB-41870: 
BG505.664 Env SOSIP in complex with polyclonal antibodies from NHP 8131 (gp120-gp120 interface epitope)
Method: single particle / : Zhang S, Ward AB

EMDB-41871: 
BG505.664 Env SOSIP in complex with polyclonal antibodies from NHP 8147 (C3/V5, V1/V2/V3 apex, gp41 glycan hole/fusion peptide and trimer base epitopes)
Method: single particle / : Zhang S, Ward AB

EMDB-41872: 
BG505.664 Env SOSIP in complex with polyclonal antibodies from NHP 8147 (gp120 glycan hole epitope)
Method: single particle / : Zhang S, Ward AB

EMDB-41972: 
GT1.1 SOSIP in complex with wk39 polyclonal antibodies from NHP A12N030 (CD4bs, C3V5 and base epitopes)
Method: single particle / : van Schooten J, Ozorowski G, Ward AB

EMDB-41973: 
GT1.1 SOSIP in complex with wk39 polyclonal antibodies from NHP A12N030 (gp41GH/FP and base epitopes)
Method: single particle / : van Schooten J, Ozorowski G, Ward AB

EMDB-41974: 
GT1.1 SOSIP in complex with wk39 polyclonal antibodies from NHP DC8G (gp41GH/FP and gp120GH epitopes)
Method: single particle / : van Schooten J, Ozorowski G, Ward AB

EMDB-41975: 
GT1.1 SOSIP in complex with wk39 polyclonal antibodies from NHP DC8G (gp120GH and base epitopes)
Method: single particle / : van Schooten J, Ozorowski G, Ward AB

EMDB-41976: 
GT1.1 SOSIP in complex with wk39 polyclonal antibodies from NHP DC8G (V1V2V3 and gp120GH epitopes)
Method: single particle / : van Schooten J, Ozorowski G, Ward AB

EMDB-41977: 
GT1.1 SOSIP-CC2 in complex with wk80 polyclonal antibodies from NHP 8229 (V1V2V3, C3V5, CD4bs, gp41GH/FP and base epitopes)
Method: single particle / : Sewell LM, Ozorowski G, Ward AB

EMDB-41978: 
GT1.1 SOSIP-CC2 in complex with wk80 polyclonal antibodies from NHP 8239 (gp120GH, gp41GH/FP, CD4bs, and base epitopes)
Method: single particle / : Sewell LM, Ozorowski G, Ward AB

EMDB-40796: 
BG505 GT1.1 SOSIP in complex with NHP Fabs 12C11 and RM20A3
Method: single particle / : Zhang S, Torres JL, Ozorowski G, Ward AB

EMDB-40797: 
BG505 GT1.1 SOSIP in complex with NHP Fabs 21N13, 21M20 and RM20A3
Method: single particle / : Ozorowski G, Torres JL, Zhang S, Ward AB

EMDB-40815: 
BG505 SOSIP.664 in complex with wk26 human polyclonal antibodies C3V5, V1V3, N611 and base from participant 017
Method: single particle / : Karlinsey D, Ozorowski G, Ward AB

EMDB-40816: 
BG505 SOSIP.664 in complex with wk26 human polyclonal antibodies gp41-N611/FP and base from participant 03
Method: single particle / : Karlinsey D, Ozorowski G, Ward AB

EMDB-40817: 
BG505 SOSIP.664 in complex with wk26 human polyclonal antibodies gp41-N611/FP and base from participant 07
Method: single particle / : Karlinsey D, Ozorowski G, Ward AB

EMDB-40818: 
BG505 SOSIP.664 in complex with wk26 human polyclonal antibodies gp120-GH and base from participant 09
Method: single particle / : Karlinsey D, Ozorowski G, Ward AB

EMDB-40819: 
BG505 SOSIP.664 in complex with wk26 human polyclonal antibodies C3V5, V1V3, gp41-GH/FP and base from participant 11
Method: single particle / : Karlinsey D, Ozorowski G, Ward AB

EMDB-40088: 
HIV-1 Env subtype C CZA97.12 SOSIP.664 in complex with 3BNC117 Fab
Method: single particle / : Ozorowski G, Lee JH, Ward AB

EMDB-24675: 
AMC018 SOSIP.v4.2 in complex with PGV04 Fab
Method: single particle / : Cottrell CA, de Val N, Ward AB

EMDB-24676: 
AMC016 SOSIP.v4.2 in complex with PGV04 Fab
Method: single particle / : Bader DLV, Cottrell CA, Ward AB
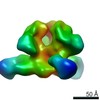
EMDB-24243: 
RM12K in complex with BG505 SOSIP
Method: single particle / : Cottrell CA, Ward AB

EMDB-24244: 
RM54B1 Fab in complex with BG505 SOSIP
Method: single particle / : Cottrell CA, Ward AB

EMDB-24245: 
RM15E Fab in complex with BG505 SOSIP
Method: single particle / : Cottrell CA, Ward AB
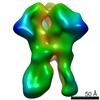
EMDB-24246: 
RM15A Fab in complex with BG505 SOSIP
Method: single particle / : Cottrell CA, Ward AB
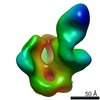
EMDB-24247: 
RM35A1 Fab in complex with BG505 SOSIP
Method: single particle / : Cottrell CA, Ward AB
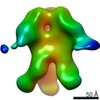
EMDB-23175: 
nsEM maps of BG505 SOSIP MD39 in complex with the polyclonal Fab samples from Group 1 of immunized rhesus macaques (Animal IDs: Rh.32034, Rh.32113, Rh.33104, Rh.33395, Rh.34909, Rh.34943)
Method: single particle / : Antanasijevic A, Sewall LM, Ward A
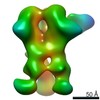
EMDB-23176: 
nsEM maps of BG505 SOSIP.v5.2 in complex with the polyclonal Fab samples from Group 2 of immunized rhesus macaques (Animal IDs: Rh.33182, Rh.33203, Rh.33311, Rh.34686, Rh.CD99, Rh.CG41)
Method: single particle / : Antanasijevic A, Sewall LM, Ward AB

EMDB-23177: 
nsEM maps of BG505 SOSIP.v5.2(7S) in complex with the polyclonal Fab samples from Group 3 of immunized rhesus macaques (Wk8 time point; Animal IDs: Rh.33172, Rh.34919, Rh.34167, Rh.33065, Rh.33176, Rh.34725)
Method: single particle / : Antanasijevic A, Sewall LM, Ward AB

EMDB-23178: 
nsEM maps of BG505 SOSIP.v5.2(7S) in complex with the polyclonal Fab samples from Group 3 of immunized rhesus macaques (Wk10 time point; Animal IDs: Rh.33172, Rh.34919, Rh.34167, Rh.33065, Rh.33176, Rh.34725)
Method: single particle / : Antanasijevic A, Sewall LM, Ward AB

EMDB-23179: 
nsEM maps of BG505 SOSIP.v5.2(7S) in complex with the polyclonal Fab samples from Group 3 of immunized rhesus macaques (Wk26 time point; Animal IDs: Rh.33172, Rh.34919, Rh.34167, Rh.33065, Rh.34725)
Method: single particle / : Antanasijevic A, Sewall LM, Ward AB

EMDB-23180: 
nsEM maps of BG505 SOSIP.v5.2(7S) in complex with the polyclonal Fab samples from Group 3 of immunized rhesus macaques (Wk38 time point; Animal IDs: Rh.33172, Rh.34919, Rh.34167, Rh.33065, Rh.34725)
Method: single particle / : Antanasijevic A, Sewall LM, Ward AB

EMDB-23181: 
nsEM map of BG505 SOSIP.v5.2(7S) in complex with the polyclonal Fab sample from animal Rh.33172 (Wk26 time point)
Method: single particle / : Antanasijevic A, Sewall LM, Ward AB

EMDB-23182: 
nsEM map of BG505 SOSIP.v3 in complex with the polyclonal Fab sample from animal Rh.33172 (Wk26 time point)
Method: single particle / : Antanasijevic A, Sewall LM, Ward AB
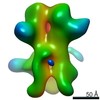
EMDB-23183: 
nsEM map of BG505 SOSIP.v3 in complex with the polyclonal Fab sample from animal Rh.33311 (Wk26 time point)
Method: single particle / : Antanasijevic A, Sewall LM, Ward AB
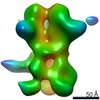
EMDB-23184: 
nsEM map of BG505 SOSIP.v5.2 in complex with the polyclonal Fab sample from animal Rh.33311 (Wk26 time point)
Method: single particle / : Antanasijevic A, Sewall LM, Ward AB
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
