[English] 日本語
 Yorodumi
Yorodumi- EMDB-23183: nsEM map of BG505 SOSIP.v3 in complex with the polyclonal Fab sam... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-23183 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | nsEM map of BG505 SOSIP.v3 in complex with the polyclonal Fab sample from animal Rh.33311 (Wk26 time point) | |||||||||
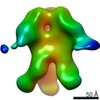
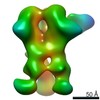
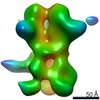
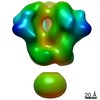
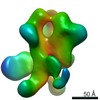
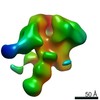
 Map data Map data | 3D map of BG505 SOSIP.v3 in complex with the polyclonal Fab sample from animal Rh.33311 (Wk26 time point, comparative analysis). Obtained by negative stain EM. | |||||||||
 Sample Sample |
| |||||||||
| Function / homology |  Function and homology information Function and homology informationsymbiont-mediated perturbation of host defense response / positive regulation of plasma membrane raft polarization / positive regulation of receptor clustering / host cell endosome membrane / clathrin-dependent endocytosis of virus by host cell / viral protein processing / fusion of virus membrane with host plasma membrane / fusion of virus membrane with host endosome membrane / viral envelope / virion attachment to host cell ...symbiont-mediated perturbation of host defense response / positive regulation of plasma membrane raft polarization / positive regulation of receptor clustering / host cell endosome membrane / clathrin-dependent endocytosis of virus by host cell / viral protein processing / fusion of virus membrane with host plasma membrane / fusion of virus membrane with host endosome membrane / viral envelope / virion attachment to host cell / host cell plasma membrane / virion membrane / structural molecule activity / identical protein binding / membrane Similarity search - Function | |||||||||
| Biological species |   Human immunodeficiency virus 1 Human immunodeficiency virus 1 | |||||||||
| Method | single particle reconstruction / negative staining / Resolution: 19.7 Å | |||||||||
 Authors Authors | Antanasijevic A / Sewall LM / Ward AB | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2021 Journal: Nat Commun / Year: 2021Title: Polyclonal antibody responses to HIV Env immunogens resolved using cryoEM. Authors: Aleksandar Antanasijevic / Leigh M Sewall / Christopher A Cottrell / Diane G Carnathan / Luis E Jimenez / Julia T Ngo / Jennifer B Silverman / Bettina Groschel / Erik Georgeson / Jinal ...Authors: Aleksandar Antanasijevic / Leigh M Sewall / Christopher A Cottrell / Diane G Carnathan / Luis E Jimenez / Julia T Ngo / Jennifer B Silverman / Bettina Groschel / Erik Georgeson / Jinal Bhiman / Raiza Bastidas / Celia LaBranche / Joel D Allen / Jeffrey Copps / Hailee R Perrett / Kimmo Rantalainen / Fabien Cannac / Yuhe R Yang / Alba Torrents de la Peña / Rebeca Froes Rocha / Zachary T Berndsen / David Baker / Neil P King / Rogier W Sanders / John P Moore / Shane Crotty / Max Crispin / David C Montefiori / Dennis R Burton / William R Schief / Guido Silvestri / Andrew B Ward /    Abstract: Engineered ectodomain trimer immunogens based on BG505 envelope glycoprotein are widely utilized as components of HIV vaccine development platforms. In this study, we used rhesus macaques to evaluate ...Engineered ectodomain trimer immunogens based on BG505 envelope glycoprotein are widely utilized as components of HIV vaccine development platforms. In this study, we used rhesus macaques to evaluate the immunogenicity of several stabilized BG505 SOSIP constructs both as free trimers and presented on a nanoparticle. We applied a cryoEM-based method for high-resolution mapping of polyclonal antibody responses elicited in immunized animals (cryoEMPEM). Mutational analysis coupled with neutralization assays were used to probe the neutralization potential at each epitope. We demonstrate that cryoEMPEM data can be used for rapid, high-resolution analysis of polyclonal antibody responses without the need for monoclonal antibody isolation. This approach allowed to resolve structurally distinct classes of antibodies that bind overlapping sites. In addition to comprehensive mapping of commonly targeted neutralizing and non-neutralizing epitopes in BG505 SOSIP immunogens, our analysis revealed that epitopes comprising engineered stabilizing mutations and of partially occupied glycosylation sites can be immunogenic. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_23183.map.gz emd_23183.map.gz | 20.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-23183-v30.xml emd-23183-v30.xml emd-23183.xml emd-23183.xml | 17.1 KB 17.1 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_23183.png emd_23183.png | 73.1 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-23183 http://ftp.pdbj.org/pub/emdb/structures/EMD-23183 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-23183 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-23183 | HTTPS FTP |
-Related structure data
| Related structure data |  7l7tC  7l7uC  7l85C  7l86C  7l87C  7l88C  7l89C  7l8aC  7l8bC  7l8cC  7l8dC  7l8eC  7l8fC  7l8gC  7l8sC  7l8tC  7l8uC  7l8wC  7l8xC  7l8yC  7l8zC  7l90C C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_23183.map.gz / Format: CCP4 / Size: 27 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_23183.map.gz / Format: CCP4 / Size: 27 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | 3D map of BG505 SOSIP.v3 in complex with the polyclonal Fab sample from animal Rh.33311 (Wk26 time point, comparative analysis). Obtained by negative stain EM. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2.05 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : BG505 SOSIP.v3 in complex with the polyclonal Fab sample from Gro...
| Entire | Name: BG505 SOSIP.v3 in complex with the polyclonal Fab sample from Group 2 of immunized rhesus macaques (Wk26 time point; Animal ID: Rh.33311) |
|---|---|
| Components |
|
-Supramolecule #1: BG505 SOSIP.v3 in complex with the polyclonal Fab sample from Gro...
| Supramolecule | Name: BG505 SOSIP.v3 in complex with the polyclonal Fab sample from Group 2 of immunized rhesus macaques (Wk26 time point; Animal ID: Rh.33311) type: complex / ID: 1 / Parent: 0 / Macromolecule list: all Details: Polyclonal complexes were generated by incubation of isolated polyclonal Fabs with recombinantly expressed BG505 SOSIP and subsequent SEC purification. |
|---|---|
| Source (natural) | Organism:   Human immunodeficiency virus 1 Human immunodeficiency virus 1 |
| Recombinant expression | Organism:  Homo sapiens (human) / Recombinant strain: HEK293F / Recombinant plasmid: pPPI4 Homo sapiens (human) / Recombinant strain: HEK293F / Recombinant plasmid: pPPI4 |
-Macromolecule #1: BG505 SOSIP.v3
| Macromolecule | Name: BG505 SOSIP.v3 / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Human immunodeficiency virus 1 Human immunodeficiency virus 1 |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MDAMKRGLCC VLLLCGAVFV SPSQEIHARF RRGARAENLW VTVYYGVPVW KDAETTLFCA SDAKAYETEK HNVWATHACV PTDPNPQEIH LENVTEEFNM WKNNMVEQMH TDIISLWDQS LKPCVKLTPL CVTLQCTNVT NNITDDMRGE LKNCSFNMTT ELRDKKQKVY ...String: MDAMKRGLCC VLLLCGAVFV SPSQEIHARF RRGARAENLW VTVYYGVPVW KDAETTLFCA SDAKAYETEK HNVWATHACV PTDPNPQEIH LENVTEEFNM WKNNMVEQMH TDIISLWDQS LKPCVKLTPL CVTLQCTNVT NNITDDMRGE LKNCSFNMTT ELRDKKQKVY SLFYRLDVVQ INENQGNRSN NSNKEYRLIN CNTSAITQAC PKVSFEPIPI HYCAPAGFAI LKCKDKKFNG TGPCPSVSTV QCTHGIKPVV STQLLLNGSL AEEEVMIRSE NITNNAKNIL VQFNTPVQIN CTRPNNNTRK SIRIGPGQAF YATGDIIGDI RQAHCNVSKA TWNETLGKVV KQLRKHFGNN TIIRFANSSG GDLEVTTHSF NCGGEFFYCN TSGLFNSTWI SNTSVQGSNS TGSNDSITLP CRIKQIINMW QRIGQAMYAP PIQGVIRCVS NITGLILTRD GGSTNSTTET FRPGGGDMRD NWRSELYKYK VVKIEPLGVA PTRCKRRVVG RRRRRRAVGI GAVFLGFLGA AGSTMGAASM TLTVQARNLL SGIVQQQSNL LRAPEAQQHL LKLTVWGIKQ LQARVLAVER YLRDQQLLGI WGCSGKLICC TNVPWNSSWS NRNLSEIWDN MTWLQWDKEI SNYTQIIYGL LEESQNQQEK NEQDLLALD |
-Experimental details
-Structure determination
| Method | negative staining |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.05 mg/mL | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.4 Component:
Details: TBS buffer, pH 7.4 | |||||||||
| Staining | Type: NEGATIVE / Material: Uranyl Formate Details: Sample diluted to 0.05 mg/mL. 3 uL was applied onto the grid, blotted off, and then stained with 2% uranyl formate for 60 seconds. | |||||||||
| Grid | Model: Homemade / Material: COPPER / Mesh: 400 / Support film - Material: CARBON / Support film - topology: CONTINUOUS / Pretreatment - Type: GLOW DISCHARGE | |||||||||
| Details | Polyclonal complexes were generated by incubation of isolated polyclonal Fabs with recombinantly expressed BG505 SOSIP and subsequent SEC purification. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI SPIRIT |
|---|---|
| Image recording | Film or detector model: TVIPS TEMCAM-F416 (4k x 4k) / Average electron dose: 25.0 e/Å2 |
| Electron beam | Acceleration voltage: 120 kV / Electron source: LAB6 |
| Electron optics | C2 aperture diameter: 100.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 1.5 µm / Nominal defocus min: 1.5 µm / Nominal magnification: 52000 |
| Sample stage | Specimen holder model: SIDE ENTRY, EUCENTRIC |
| Experimental equipment |  Model: Tecnai Spirit / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: RIGID BODY FIT |
|---|
 Movie
Movie Controller
Controller


 UCSF Chimera
UCSF Chimera






































 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)





















