-Search query
-Search result
Showing 1 - 50 of 176 items for (author: hwang & t)

EMDB-71899: 
Structure of V30V4 in complex with SARS-CoV-2 spike
Method: single particle / : Wang YJ, Kibria G, Wesemann D, Chen B

PDB-9pw4: 
Structure of V30V4 in complex with SARS-CoV-2 spike
Method: single particle / : Wang YJ, Kibria G, Wesemann D, Chen B

EMDB-71900: 
The local refinement map of Structure of V30V4 in complex with SARS-CoV-2 spike
Method: single particle / : Wang YJ, Kibria G, Wesemann D, Chen B

EMDB-48538: 
CryoEM Structure of the Candida albicans Group I Intron-GMP Complex
Method: single particle / : Chung K, Xu L, Liu T, Pyle A

EMDB-48539: 
CryoEM Structure of the Candida albicans Group I Intron-Compound 11 Complex under Magnesium Condition
Method: single particle / : Chung K, Xu L, Liu T, Pyle A

EMDB-48540: 
CryoEM Structure of the Candida albicans Group I Intron-Compound 11 Complex under Calcium Condition
Method: single particle / : Chung K, Xu L, Liu T, Pyle A

PDB-9mqs: 
CryoEM Structure of the Candida albicans Group I Intron-GMP Complex
Method: single particle / : Chung K, Xu L, Liu T, Pyle A

PDB-9mqt: 
CryoEM Structure of the Candida albicans Group I Intron-Compound 11 Complex under Magnesium Condition
Method: single particle / : Chung K, Xu L, Liu T, Pyle A

PDB-9mqu: 
CryoEM Structure of the Candida albicans Group I Intron-Compound 11 Complex under Calcium Condition
Method: single particle / : Chung K, Xu L, Liu T, Pyle A

EMDB-60459: 
AtALMT9 with LMNG and sterol mimic CHS (sterol1 class)
Method: single particle / : Lee Y, Lee S

EMDB-60461: 
AtALMT9 with LMNG and sterol mimic CHS (sterol2 class)
Method: single particle / : Lee Y, Lee S

EMDB-60462: 
AtALMT9 with LMNG (narrow class)
Method: single particle / : Lee Y, Lee S

EMDB-60463: 
AtALMT9 with LMNG (wide class)
Method: single particle / : Lee Y, Lee S

EMDB-60464: 
AtALMT9 with LMNG (cis1-PI4P class)
Method: single particle / : Lee Y, Lee S

EMDB-60465: 
AtALMT9 with LMNG (cis2 class)
Method: single particle / : Lee Y, Lee S

EMDB-60466: 
AtALMT9 with LMNG (intermediate class)
Method: single particle / : Lee Y, Lee S

EMDB-60467: 
AtALMT9 with LMNG (trans1 class)
Method: single particle / : Lee Y, Lee S

EMDB-60468: 
AtALMT9 with LMNG (trans2 class)
Method: single particle / : Lee Y, Lee S

EMDB-61818: 
AtALMT1 with LMNG and sterol mimic CHS
Method: single particle / : Lee Y, Lee S

PDB-8zte: 
AtALMT9 with LMNG and sterol mimic CHS (sterol1 class)
Method: single particle / : Lee Y, Lee S

PDB-8ztg: 
AtALMT9 with LMNG and sterol mimic CHS (sterol2 class)
Method: single particle / : Lee Y, Lee S

EMDB-45467: 
Kalium channelrhodopsin 1 C110A mutant from Hyphochytrium catenoides, Dark State
Method: single particle / : Morizumi T, Kim K, Ernst OP

EMDB-45468: 
Kalium channelrhodopsin 1 C110A mutant from Hyphochytrium catenoides, Laser-Flash-Illuminated
Method: single particle / : Morizumi T, Kim K, Ernst OP

EMDB-45469: 
Kalium channelrhodopsin 1 C110A mutant from Hyphochytrium catenoides, Continuous Illumination State
Method: single particle / : Morizumi T, Kim K, Ernst OP

PDB-9cdc: 
Kalium channelrhodopsin 1 C110A mutant from Hyphochytrium catenoides, Dark State
Method: single particle / : Morizumi T, Kim K, Ernst OP

PDB-9cdd: 
Kalium channelrhodopsin 1 C110A mutant from Hyphochytrium catenoides, Laser-Flash-Illuminated
Method: single particle / : Morizumi T, Kim K, Ernst OP

PDB-9cde: 
Kalium channelrhodopsin 1 C110A mutant from Hyphochytrium catenoides, Continuous Illumination State
Method: single particle / : Morizumi T, Kim K, Ernst OP

EMDB-45360: 
Cryo-EM structure of TAP binding protein related (TAPBPR) in complex with HLA-A*02:01 bound to a suboptimal peptide.
Method: single particle / : Pumroy RP, Mallik L, Sun Y, Moiseenkova-Bell YV, Sgourakis NG

PDB-9c96: 
Cryo-EM structure of TAP binding protein related (TAPBPR) in complex with HLA-A*02:01 bound to a suboptimal peptide.
Method: single particle / : Pumroy RP, Mallik L, Sun Y, Moiseenkova-Bell YV, Sgourakis NG
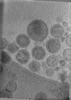
EMDB-47705: 
Tomogram of Cas9 containing Enveloped Delivery Vehicles (EDVs)
Method: electron tomography / : Peukes J, Ngo W, Nogales E, Doudna J
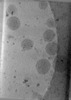
EMDB-47741: 
Tomogram of Enveloped Delivery Vehicles (EDVs) without Cas9
Method: electron tomography / : Peukes J, Ngo W, Nogales E, Doudna J

EMDB-47743: 
Subtomogram average of capsid (CA) from immature Enveloped Delivery Vehicles (EDVs) without Cas9
Method: subtomogram averaging / : Peukes J, Ngo W, Nogales E, Doudna J

EMDB-47745: 
Subtomogram average of capsid (CA) from immature Enveloped Delivery Vehicles (EDVs) containing Cas9
Method: subtomogram averaging / : Peukes J, Ngo W, Nogales E, Doudna J
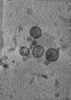
EMDB-47858: 
Tomogram of a minimized Enveloped Delivery Vehicle (miniEDV)
Method: electron tomography / : Peukes J, Ngo W, Nogales E, Doudna J

EMDB-41346: 
CRYO-EM STRUCTURE OF HIV-1 BG505DS-SOSIP.664 ENV TRIMER BOUND TO DJ85-b.01 FAB
Method: single particle / : Pletnev S, Hoyt F, Fischer E, Kwong P

EMDB-41359: 
CRYO-EM STRUCTURE OF HIV-1 BG505DS-SOSIP.664 ENV TRIMER BOUND TO DJ85-c.01 FAB
Method: single particle / : Pletnev S, Hoyt F, Fischer E, Kwong P

EMDB-41360: 
CRYO-EM STRUCTURE OF HIV-1 BG505DS-SOSIP.664 ENV TRIMER BOUND TO DJ85-d.01 FAB
Method: single particle / : Pletnev S, Hoyt F, Fischer E, Kwong P

EMDB-41361: 
CRYO-EM STRUCTURE OF HIV-1 BG505DS-SOSIP.664 ENV TRIMER BOUND TO DJ85-e.01 FAB
Method: single particle / : Pletnev S, Hoyt F, Fischer E, Kwong P

EMDB-41362: 
CRYO-EM STRUCTURE OF HIV-1 BG505DS-SOSIP.664 ENV TRIMER BOUND TO HERH-c.01 FAB
Method: single particle / : Pletnev S, Hoyt F, Fischer E, Kwong P

PDB-8tkc: 
CRYO-EM STRUCTURE OF HIV-1 BG505DS-SOSIP.664 ENV TRIMER BOUND TO DJ85-b.01 FAB
Method: single particle / : Pletnev S, Hoyt F, Fischer E, Kwong P

PDB-8tl2: 
CRYO-EM STRUCTURE OF HIV-1 BG505DS-SOSIP.664 ENV TRIMER BOUND TO DJ85-c.01 FAB
Method: single particle / : Pletnev S, Hoyt F, Fischer E, Kwong P

PDB-8tl3: 
CRYO-EM STRUCTURE OF HIV-1 BG505DS-SOSIP.664 ENV TRIMER BOUND TO DJ85-d.01 FAB
Method: single particle / : Pletnev S, Hoyt F, Fischer E, Kwong P
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search











 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
