-Search query
-Search result
Showing all 33 items for (author: hsu & hc)

EMDB-40846: 
The C-terminal protease CtpA-LbcA complex of pseudomonas aeruginosa with the TPR at the low position

EMDB-40847: 
The C-terminal protease CtpA-LbcA complex of pseudomonas aeruginosa with the TPR at the high position

EMDB-40848: 
The structure of C-terminal protease CtpA-LbcA complex of Pseudomonas aeruginosa

EMDB-40849: 
Structure of the C-terminal protease CtpA-LbcA complex of Pseudomonas aeruginosa

EMDB-40850: 
The C-terminal protease CtpA-LbcA complex of pseudomonas aeruginosa with the TPR at the high position
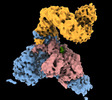
EMDB-40851: 
The C-terminal protease CtpA-LbcA complex of pseudomonas aeruginosa with the TPR at the low position

EMDB-40852: 
Structure of the C-terminal protease CtpA-LbcA complex of Pseudomonas aeruginosa
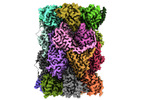
EMDB-29764: 
Structure of the Plasmodium falciparum 20S proteasome complexed with inhibitor TDI-8304
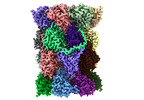
EMDB-29765: 
Structure of the Plasmodium falciparum 20S proteasome beta-6 A117D mutant complexed with inhibitor WLW-vs

EMDB-42148: 
Structure of human constitutive 20S proteasome complexed with the inhibitor TDI-8304

EMDB-29603: 
The PI31-free Bovine 20S proteasome

EMDB-29604: 
The human PI31 complexed with bovine 20S proteasome

EMDB-31820: 
Negative staining (NS)-EM structure of SARS-CoV-2 S-Kappa variant in complex with neutralizing antibodies, RBD-chAb-15 and RBD-chAb45
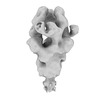
EMDB-31818: 
Negative staining (NS)-EM structure of SARS-CoV-2 S-Gamma variant in complex with neutralizing antibodies, RBD-chAb-15 and RBD-chAb45

EMDB-31822: 
Negative staining (NS)-EM structure of SARS-CoV-2 S-Kappa variant in complex with neutralizing antibody RBD-chAb-25

EMDB-31821: 
Negative staining (NS)-EM structure of SARS-CoV-2 S-Delta variant in complex with neutralizing antibody RBD-chAb-25

EMDB-31817: 
Negative staining (NS)-EM structure of SARS-CoV-2 S-Beta variant in complex with neutralizing antibodies, RBD-chAb-15 and RBD-chAb45

EMDB-31819: 
Negative staining (NS)-EM structure of SARS-CoV-2 S-Delta variant in complex with neutralizing antibodies, RBD-chAb-15 and RBD-chAb45

EMDB-32832: 
SARS-CoV-2 Spike in complex with Fab of m31A7

EMDB-32825: 
Negative stain volume of the mono-GlcNAc-decorated SARS-CoV-2 Spike

EMDB-31069: 
Cryo-EM structure of SARS-CoV-2 S-UK variant (B.1.1.7), one RBD-up conformation 1

EMDB-31070: 
Cryo-EM structure of SARS-CoV-2 S-UK variant (B.1.1.7), one RBD-up conformation 2

EMDB-31071: 
Cryo-EM structure of SARS-CoV-2 S-UK variant (B.1.1.7), one RBD-up conformation 3

EMDB-31072: 
Cryo-EM structure of SARS-CoV-2 S-UK variant (B.1.1.7), two RBD-up conformation

EMDB-31073: 
Cryo-EM structure of SARS-CoV-2 S-UK variant (B.1.1.7) in complex with Angiotensin-converting enzyme 2 (ACE2) ectodomain

EMDB-31074: 
Cryo-EM structure of SARS-CoV-2 S-D614G variant in complex with neutralizing antibodies, RBD-chAb-15 and RBD-chAb45

EMDB-31470: 
Cryo-EM structure of SARS-CoV-2 spike in complex with a neutralizing antibody chAb-25 (Focused refinement of S-RBD and chAb-25 region)

EMDB-31471: 
Cryo-EM structure of SARS-CoV-2 spike in complex with a neutralizing antibody chAb-45 (Focused refinement of S-RBD and chAb-45 region)

EMDB-31473: 
Cryo-EM structure of SARS-CoV-2 spike in complex with two neutralizing antibodies chAb-25 and chAb-45

EMDB-30669: 
Cryo-EM structure of SARS-CoV-2 spike in complex with a neutralizing antibody chAb-25

EMDB-30670: 
Cryo-EM structure of SARS-CoV-2 spike in complex with a neutralizing antibody chAb-45

EMDB-20467: 
Cryo-EM structure of S. cerevisiae Drs2p-Cdc50p in the PI4P-activated form

EMDB-20468: 
Cryo-EM structure of S. cerevisiae Drs2p-Cdc50p in the autoinhibited apo form
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
