-Search query
-Search result
Showing 1 - 50 of 427 items for (author: francis & f)

EMDB-19075: 
Conformational Landscape of the Type V-K CRISPR-associated TransposonIntegration Assembly CAST V-K composite map

EMDB-19282: 
Conformational Landscape of the Type V-K CRISPR-associated TransposonIntegration Assembly CAST V-K Cas12k domain local-refinement map

EMDB-19283: 
Conformational Landscape of the Type V-K CRISPR-associated TransposonIntegration Assembly CAST V-K TnsC domain local-refinement map

EMDB-19284: 
Conformational Landscape of the Type V-K CRISPR-associated TransposonIntegration Assembly CAST V-K TnsB domain local-refinement map

EMDB-19286: 
consensus map of the V-K CRISPR-associated Transposon Integration Assembly

PDB-8rdu: 
Conformational Landscape of the Type V-K CRISPR-associated TransposonIntegration Assembly CAST V-K composite map

PDB-8rkt: 
Conformational Landscape of the Type V-K CRISPR-associated TransposonIntegration Assembly CAST V-K Cas12k domain local-refinement map

PDB-8rku: 
Conformational Landscape of the Type V-K CRISPR-associated TransposonIntegration Assembly CAST V-K TnsC domain local-refinement map

PDB-8rkv: 
Conformational Landscape of the Type V-K CRISPR-associated TransposonIntegration Assembly CAST V-K TnsB domain local-refinement map

EMDB-15697: 
Cryo-EM structure of shCas12k-sgRNA-dsDNA ternary complex (type V-K CRISPR-associated transposon)

PDB-8axa: 
Cryo-EM structure of shCas12k-sgRNA-dsDNA ternary complex (type V-K CRISPR-associated transposon)
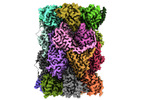
EMDB-29764: 
Structure of the Plasmodium falciparum 20S proteasome complexed with inhibitor TDI-8304
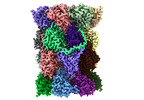
EMDB-29765: 
Structure of the Plasmodium falciparum 20S proteasome beta-6 A117D mutant complexed with inhibitor WLW-vs

EMDB-42148: 
Structure of human constitutive 20S proteasome complexed with the inhibitor TDI-8304

PDB-8g6e: 
Structure of the Plasmodium falciparum 20S proteasome complexed with inhibitor TDI-8304

PDB-8g6f: 
Structure of the Plasmodium falciparum 20S proteasome beta-6 A117D mutant complexed with inhibitor WLW-vs

PDB-8ud9: 
Structure of human constitutive 20S proteasome complexed with the inhibitor TDI-8304

EMDB-16512: 
MiniCoV-ADDomer, a SARS-CoV-2 epitope presenting viral like particle

EMDB-16522: 
Structure of ADDoCoV-ADAH11

PDB-8c9n: 
MiniCoV-ADDomer, a SARS-CoV-2 epitope presenting viral like particle

EMDB-17819: 
XBB 1.0 RBD bound to P4J15 (Local)
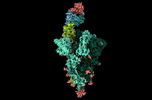
EMDB-17849: 
XBB 1.0 RBD bound to P4J15 (Global)

EMDB-17850: 
SARS-CoV-2 XBB 1.0 closed conformation.

PDB-8pq2: 
XBB 1.0 RBD bound to P4J15 (Local)

PDB-8psd: 
SARS-CoV-2 XBB 1.0 closed conformation.

EMDB-41144: 
Cryo-EM Structure of GPR61-G protein complex stabilized by scFv16

EMDB-41145: 
Cryo-EM Structure of GPR61-

PDB-8tb0: 
Cryo-EM Structure of GPR61-G protein complex stabilized by scFv16

PDB-8tb7: 
Cryo-EM Structure of GPR61-

EMDB-16521: 
Cryo-EM structure of the Cibeles-Demetra 3:3 heterocomplex from Galleria mellonella saliva

EMDB-16524: 
Cryo-EM structure of the Ceres homohexamer from Galleria mellonella saliva

EMDB-16531: 
Cryo-EM structure of the Cora homohexamer from Galleria mellonella saliva

PDB-8ca9: 
Cryo-EM structure of the Cibeles-Demetra 3:3 heterocomplex from Galleria mellonella saliva

PDB-8cad: 
Cryo-EM structure of the Ceres homohexamer from Galleria mellonella saliva

PDB-8can: 
Cryo-EM structure of the Cora homohexamer from Galleria mellonella saliva

EMDB-28378: 
Structure of the C3bB proconvertase in complex with lufaxin and factor Xa

PDB-8eok: 
Structure of the C3bB proconvertase in complex with lufaxin and factor Xa

EMDB-28279: 
Structure of the C3bB proconvertase in complex with lufaxin

PDB-8enu: 
Structure of the C3bB proconvertase in complex with lufaxin

EMDB-28228: 
SARS-CoV-2 spike glycoprotein in complex with the ICO-hu23 neutralizing antibody Fab fragment

PDB-8elj: 
SARS-CoV-2 spike glycoprotein in complex with the ICO-hu23 neutralizing antibody Fab fragment

EMDB-29930: 
T. cruzi topoisomerase II alpha bound to dsDNA and the covalent inhibitor CT1

PDB-8gcc: 
T. cruzi topoisomerase II alpha bound to dsDNA and the covalent inhibitor CT1

EMDB-16900: 
Cryo-EM reconstruction of the native 24-mer E2o core of the 2-oxoglutarate dehydrogenase complex of C. thermophilum at 3.35 A resolution

PDB-8oiu: 
Cryo-EM reconstruction of the native 24-mer E2o core of the 2-oxoglutarate dehydrogenase complex of C. thermophilum at 3.35 A resolution

EMDB-40422: 
Cryo-EM Structure of RyR1

EMDB-40423: 
Cryo-EM Structure of RyR1 + ATP-gamma-S

EMDB-40424: 
Cryo-EM Structure of RyR1 + ADP

EMDB-40425: 
Cryo-EM Structure of RyR1 + AMP

EMDB-40426: 
Cryo-EM Structure of RyR1 + Adenosine
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
