-Search query
-Search result
Showing all 49 items for (author: davey & r)

EMDB-14986: 
CryoEM structure of Ku heterodimer bound to DNA

PDB-7zvt: 
CryoEM structure of Ku heterodimer bound to DNA

EMDB-14955: 
Cryo-EM structure of Ku 70/80 bound to inositol hexakisphosphate

PDB-7zt6: 
Cryo-EM structure of Ku 70/80 bound to inositol hexakisphosphate

EMDB-26005: 
Structure of the Inmazeb cocktail and resistance to escape against Ebola virus

PDB-7tn9: 
Structure of the Inmazeb cocktail and resistance to escape against Ebola virus

EMDB-26372: 
SARS-2 CoV 6P Mut7 in complex with Fab CC84.5

EMDB-26522: 
SARS-CoV-2 6P Mut7 in complex with K398.25 Fab

EMDB-26523: 
SARS-CoV-1 in complex with K398.25 Fab

EMDB-26524: 
SARS-CoV-2 6P Mut7 in complex with K398.16 Fab
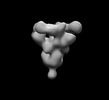
EMDB-26525: 
SARS-CoV-2 6P Mut7 in complex with K398.16 Fab (3 bound)

EMDB-26526: 
SARS-CoV-1 in complex with K398.16 Fab

EMDB-26527: 
SARS-CoV-2 6P Mut7 in complex with K288.2 Fab

EMDB-26528: 
SARS-CoV-2 6P Mut7 in complex with K398.8 Fab

EMDB-26529: 
SARS-CoV-2 6P Mut7 in complex with K398.8 Fab (2 bound)

EMDB-26530: 
SARS-CoV-2 6P Mut7 in complex with K398.18 Fab

EMDB-26531: 
SARS-CoV-2 6P Mut7 in complex with K398.18 Fabs (2 bound)

EMDB-26532: 
SARS-CoV-2 6P Mut7 in complex with K398.22 Fab
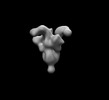
EMDB-26533: 
SARS-CoV-2 6P Mut7 in complex with K398.22 Fab (2 bound)

EMDB-26534: 
SARS-CoV-1 in complex with K398.8 Fab

EMDB-26535: 
SARS-CoV-1 in complex with K398.8 Fab (2 bound)

EMDB-26536: 
SARS-CoV-1 in complex with K398.18 Fab (2 bound)

EMDB-26537: 
SARS-CoV-1 in complex with K398.18 Fab (3 bound)

EMDB-26538: 
SARS-CoV-1 in complex with K288.2 Fab

EMDB-26539: 
SARS-CoV-1 in complex with K398.22 Fab

EMDB-26365: 
SARS-2 CoV 6P Mut7 in complex with Fab CC25.1
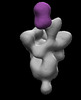
EMDB-26366: 
SARS-2 CoV 6P Mut7 in complex with Fab CC25.54

EMDB-26367: 
SARS-2 CoV 6P Mut7 + Fab CC25.52

EMDB-26368: 
SARS-2 CoV 6P Mut7 in complex with Fab CC25.56
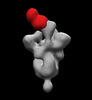
EMDB-26369: 
SARS-2 CoV 6P Mut7 in complex with Fab CC25.36

EMDB-26370: 
SARS-2 CoV 6P Mut7 in complex with Fab CC25.3
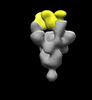
EMDB-26371: 
SARS-2 CoV 6P Mut7 in complex with Fab CC25.11

EMDB-26373: 
SARS-2 CoV 6P Mut7 in complex with Fab CC84.2

EMDB-26374: 
SARS-2 CoV 6P Mut7 in complex with Fab CC84.10
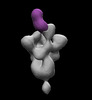
EMDB-26375: 
SARS-2 CoV 6P Mut7 in complex with Fab CC84.24

EMDB-7900: 
REGN3479 antibody Fab in complex with Ebola virus GP

EMDB-7901: 
REGN3470 antibody Fab in complex with Ebola virus GP

EMDB-7902: 
REGN3471 antibody Fab in complex with Ebola virus GP

EMDB-7089: 
HIV-1 Envelope SOSIP trimer clone PC64M4c054 in complex with autologous PCT64-13C Fab at 13.2 A resolution

EMDB-7104: 
HIV-1 Envelope Glycoprotein clone PC64M4c054 SOSIP

EMDB-7105: 
HIV-1 Envelope Glycoprotein clone PC64M7c022 SOSIP.664

EMDB-7106: 
HIV-1 Envelope Glycoprotein clone PC64M18c043 SOSIP.664

EMDB-7107: 
HIV-1 Envelope Glycoprotein clone PC64M4c054 SOSIP in complex with PGV04 Fab

EMDB-7108: 
HIV-1 Envelope Glycoprotein clone PC64M18c043 SOSIP.664

PDB-4ard: 
Structure of the immature retroviral capsid at 8A resolution by cryo- electron microscopy

PDB-4arg: 
Structure of the immature retroviral capsid at 8A resolution by cryo- electron microscopy

EMDB-2089: 
Structure of the immature retroviral capsid at 8A resolution by cryo-electron microscopy

EMDB-2090: 
Structure of the immature retroviral capsid at 8A resolution by cryo-electron microscopy

EMDB-1986: 
CryoEM reconstruction of the Marburg virus nucleocapsid.
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
