[English] 日本語
 Yorodumi
Yorodumi- EMDB-7107: HIV-1 Envelope Glycoprotein clone PC64M4c054 SOSIP in complex wit... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-7107 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | HIV-1 Envelope Glycoprotein clone PC64M4c054 SOSIP in complex with PGV04 Fab | |||||||||
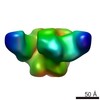
 Map data Map data | primary map | |||||||||
 Sample Sample |
| |||||||||
| Biological species |   Human immunodeficiency virus 1 / Human immunodeficiency virus 1 /  Homo sapiens (human) Homo sapiens (human) | |||||||||
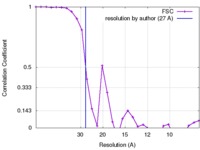
| Method | single particle reconstruction / negative staining / Resolution: 27.0 Å | |||||||||
 Authors Authors | Berndsen ZT / Rantalainen K / Ward AB | |||||||||
 Citation Citation |  Journal: Immunity / Year: 2017 Journal: Immunity / Year: 2017Title: HIV Envelope Glycoform Heterogeneity and Localized Diversity Govern the Initiation and Maturation of a V2 Apex Broadly Neutralizing Antibody Lineage. Authors: Elise Landais / Ben Murrell / Bryan Briney / Sasha Murrell / Kimmo Rantalainen / Zachary T Berndsen / Alejandra Ramos / Lalinda Wickramasinghe / Melissa Laird Smith / Kemal Eren / Natalia de ...Authors: Elise Landais / Ben Murrell / Bryan Briney / Sasha Murrell / Kimmo Rantalainen / Zachary T Berndsen / Alejandra Ramos / Lalinda Wickramasinghe / Melissa Laird Smith / Kemal Eren / Natalia de Val / Mengyu Wu / Audrey Cappelletti / Jeffrey Umotoy / Yolanda Lie / Terri Wrin / Paul Algate / Po-Ying Chan-Hui / Etienne Karita / / / Andrew B Ward / Ian A Wilson / Dennis R Burton / Davey Smith / Sergei L Kosakovsky Pond / Pascal Poignard /    Abstract: Understanding how broadly neutralizing antibodies (bnAbs) to HIV envelope (Env) develop during natural infection can help guide the rational design of an HIV vaccine. Here, we described a bnAb ...Understanding how broadly neutralizing antibodies (bnAbs) to HIV envelope (Env) develop during natural infection can help guide the rational design of an HIV vaccine. Here, we described a bnAb lineage targeting the Env V2 apex and the Ab-Env co-evolution that led to development of neutralization breadth. The lineage Abs bore an anionic heavy chain complementarity-determining region 3 (CDRH3) of 25 amino acids, among the shortest known for this class of Abs, and achieved breadth with only 10% nucleotide somatic hypermutation and no insertions or deletions. The data suggested a role for Env glycoform heterogeneity in the activation of the lineage germline B cell. Finally, we showed that localized diversity at key V2 epitope residues drove bnAb maturation toward breadth, mirroring the Env evolution pattern described for another donor who developed V2-apex targeting bnAbs. Overall, these findings suggest potential strategies for vaccine approaches based on germline-targeting and serial immunogen design. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_7107.map.gz emd_7107.map.gz | 728.8 KB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-7107-v30.xml emd-7107-v30.xml emd-7107.xml emd-7107.xml | 18.1 KB 18.1 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_7107_fsc.xml emd_7107_fsc.xml | 2.8 KB | Display |  FSC data file FSC data file |
| Images |  emd_7107.png emd_7107.png | 62.2 KB | ||
| Others |  emd_7107_half_map_1.map.gz emd_7107_half_map_1.map.gz emd_7107_half_map_2.map.gz emd_7107_half_map_2.map.gz | 736.3 KB 735.7 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-7107 http://ftp.pdbj.org/pub/emdb/structures/EMD-7107 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-7107 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-7107 | HTTPS FTP |
-Related structure data
| Related structure data |  7089C  7104C  7105C  7106C  7108C  5fehC C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_7107.map.gz / Format: CCP4 / Size: 1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_7107.map.gz / Format: CCP4 / Size: 1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | primary map | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 4.1 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Half map: half map 1
| File | emd_7107_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: half map 2
| File | emd_7107_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : HIV-1 Envelope Glycoprotein clone PC64M4c054 SOSIP.664 in complex...
| Entire | Name: HIV-1 Envelope Glycoprotein clone PC64M4c054 SOSIP.664 in complex with PGV04 Fab |
|---|---|
| Components |
|
-Supramolecule #1: HIV-1 Envelope Glycoprotein clone PC64M4c054 SOSIP.664 in complex...
| Supramolecule | Name: HIV-1 Envelope Glycoprotein clone PC64M4c054 SOSIP.664 in complex with PGV04 Fab type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:   Human immunodeficiency virus 1 Human immunodeficiency virus 1 |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 225 KDa |
-Macromolecule #1: HIV-1 Envelope Glycoprotein clone PC64M4c054 SOSIP.664
| Macromolecule | Name: HIV-1 Envelope Glycoprotein clone PC64M4c054 SOSIP.664 type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Human immunodeficiency virus 1 Human immunodeficiency virus 1 |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MDAMKRGLCC VLLLCGAVFV SPSQEIHARF RRGAR ANNL WVTVYYGVPV WRDAETTLFC ASDAKAYDTE VHNVWATHAC VPTDPSPQEI HLANVTEKFN MWKNSMVEQM HTDIISLWDE SLKPCVKLTP LCITLNCTNI TRKNVTGGNL TEDGKEELKN CSFNATTELR ...String: MDAMKRGLCC VLLLCGAVFV SPSQEIHARF RRGAR ANNL WVTVYYGVPV WRDAETTLFC ASDAKAYDTE VHNVWATHAC VPTDPSPQEI HLANVTEKFN MWKNSMVEQM HTDIISLWDE SLKPCVKLTP LCITLNCTNI TRKNVTGGNL TEDGKEELKN CSFNATTELR NKRQKVHSLF YRLDLVELNE GNSSNSNTSM YRLINCNTSA ITQACPKVSF EPIPIHYCAP AGFAILKCRE EEFNGTGPCK NVSTVQCTHG IKPVVSTQLL LNGSLAEGTV KIRCENISNN AKTILVQLTT PVRINCTRPN NNTRTSIRIG PGQSFYATGD IIGDIRKAYC NVSGSEWKEA LGKVVVQLRS HFNKTITFAS SSGGDLEITT HSFNCGGEFF YCNTSSLFNS TWDGNSTTNS TQEPNGTITL PCRIKQIINM WQRTGQAMYA PPIPGKIRCD SNITGLILTR DGENNNTESE TFRPEGGDMR NNWRSELYKY KVVKIDPLGV APTGCKRRVV ERRRRRRAVG IGAVLFGFLG AAGSTMGAAS LTLTVQARQL LSGIVQQQSN LLRAPEAQQH LLRLTVWGIK QLQARVLAVE RYLSDQQLLG IWGCSGKLIC CTTVPWNSSW SNKSQDEIWN NMTWLQWDKE ISNYTDTIYY LIEKSQNQQE VNEKDLLALD |
-Macromolecule #2: Human IgG Fab PGV04 Heavy Chain
| Macromolecule | Name: Human IgG Fab PGV04 Heavy Chain / type: protein_or_peptide / ID: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: qvqlvqsgsg vkkpgasvr v scwtsedi fe rtelihw vrq apgqgl ewig wvktv tgavn fgsp dfrqrv slt rdrdlft ah mdirgltq g dtatyfcar qkfytggqgw yfdlwgrgt l ivvssast kg psvfpla pss kstsgg taal gclvk ...String: qvqlvqsgsg vkkpgasvr v scwtsedi fe rtelihw vrq apgqgl ewig wvktv tgavn fgsp dfrqrv slt rdrdlft ah mdirgltq g dtatyfcar qkfytggqgw yfdlwgrgt l ivvssast kg psvfpla pss kstsgg taal gclvk dyfpe pvtv swnsga lts gvhtfpa vl qssglysl s svvtvpsss lgtqtyicnv nhkpsntkv d kkvepksc |
-Macromolecule #3: Human IgG Fab PGV04 Heavy Chain
| Macromolecule | Name: Human IgG Fab PGV04 Heavy Chain / type: protein_or_peptide / ID: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: eivltqspgt lslspgeta s lsctaasy gh mtwyqkk pgq ppklli fats krasg ipdrf sgsq fgkqyt lti trmeped fa ryycqqle f fgqgtrlei rrtvaapsvf ifppsdeql k sgtasvvc ll nnfypre akv qwkvdn alqs gnsqe ...String: eivltqspgt lslspgeta s lsctaasy gh mtwyqkk pgq ppklli fats krasg ipdrf sgsq fgkqyt lti trmeped fa ryycqqle f fgqgtrlei rrtvaapsvf ifppsdeql k sgtasvvc ll nnfypre akv qwkvdn alqs gnsqe svteq dskd stysls stl tlskady ek hkvyacev t hqglsspvt ksfnrgec |
-Experimental details
-Structure determination
| Method | negative staining |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Staining | Type: NEGATIVE / Material: NanoW |
| Grid | Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Details: unspecified |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI SPIRIT |
|---|---|
| Image recording | Film or detector model: TVIPS TEMCAM-F416 (4k x 4k) / Average exposure time: 0.5 sec. / Average electron dose: 25.0 e/Å2 |
| Electron beam | Acceleration voltage: 120 kV / Electron source: LAB6 |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Tecnai Spirit / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller


 UCSF Chimera
UCSF Chimera






 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)